6U0M
 
 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
1AXC
 
 | | HUMAN PCNA | | Descriptor: | P21/WAF1, PCNA | | Authors: | Gulbis, J.M, Kuriyan, J. | | Deposit date: | 1997-10-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA.
Cell(Cambridge,Mass.), 87, 1996
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTJ
 
 | | Structure of Ctf4 trimer in complex with one CMG helicase | | Descriptor: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTN
 
 | | Structure of Ctf4 trimer in complex with two CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
3PWE
 
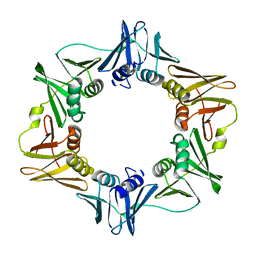 | | Crystal structure of the E. coli beta clamp mutant R103C, I305C, C260S, C333S at 2.2A resolution | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Marzahn, M.R, Robbins, A.H, McKenna, R, Bloom, L.B. | | Deposit date: | 2010-12-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The E. coli clamp loader can actively pry open the beta-sliding clamp
J.Biol.Chem., 286, 2011
|
|
8FOH
 
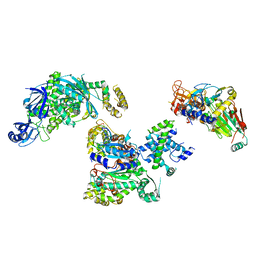 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the RNA synthesis state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOJ
 
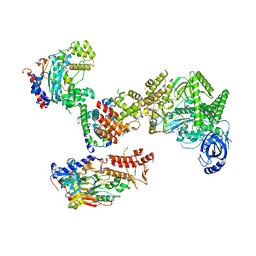 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the post RNA handoff state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOD
 
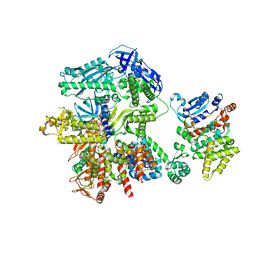 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in Apo state conformation II | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOE
 
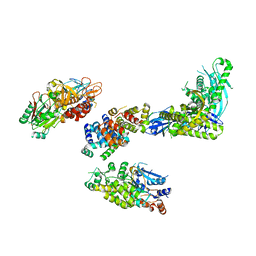 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex bound to a template DNA | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOC
 
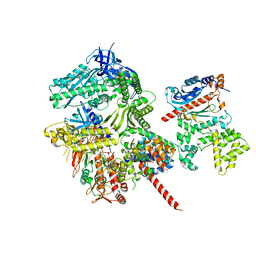 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase in Apo state conformation I | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOK
 
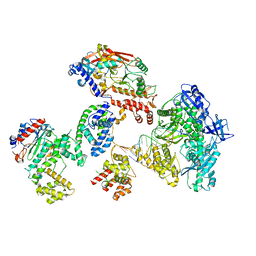 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the DNA elongation state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8UK9
 
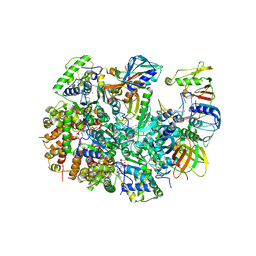 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UH7
 
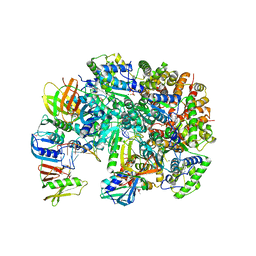 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6WJV
 
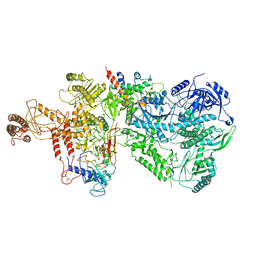 | | Structure of the Saccharomyces cerevisiae polymerase epsilon holoenzyme | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, DNA polymerase epsilon subunit C, ... | | Authors: | Yuan, Z, Georgescu, R, Schauer, G.D, O'Donnell, M, Li, H. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome.
Nat Commun, 11, 2020
|
|
2POL
 
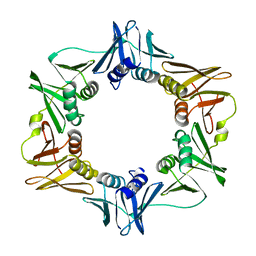 | |
1PLR
 
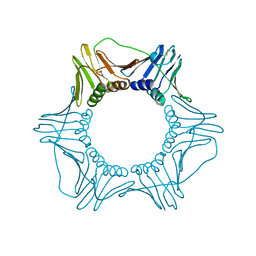 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1PLQ
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | MERCURY (II) ION, PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
3GLI
 
 | | Crystal Structure of the E. coli clamp loader bound to Primer-Template DNA and Psi Peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Cantor, A.J, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GLH
 
 | | Crystal Structure of the E. coli clamp loader bound to Psi Peptide | | Descriptor: | DNA polymerase III subunit delta, DNA polymerase III subunit delta', DNA polymerase III subunit tau | | Authors: | Kazmirski, S.L, Simonetta, K.R, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.891 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GLG
 
 | | Crystal Structure of a Mutant (gammaT157A) E. coli Clamp Loader Bound to Primer-Template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Seyedin, S.N, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GLF
 
 | |
3K4X
 
 | | Eukaryotic Sliding Clamp PCNA Bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), Proliferating cell nuclear antigen | | Authors: | McNally, R, Kuriyan, J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Analysis of the role of PCNA-DNA contacts during clamp loading.
Bmc Struct.Biol., 10, 2010
|
|
1OK7
 
 | | A Conserved protein binding-site on Bacterial Sliding Clamps | | Descriptor: | DNA POLYMERASE III, DNA POLYMERASE IV | | Authors: | Burnouf, D.Y, Olieric, V, Wagner, J, Fujii, S, Reinbolt, J, Fuchs, R.P.P, Dumas, P. | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of Sliding Clamp/Ligand Interactions Suggest a Competition between Replicative and Translesion DNA Polymerases
J.Mol.Biol., 335, 2004
|
|
