8D9H
 
 | | gRAMP-TPR-CHAT match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, PHOSPHATE ION, RAMP superfamily protein, ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
4ES3
 
 | |
4ES2
 
 | |
4QQZ
 
 | | Crystal structure of T. fusca Cas3-AMPPNP | | Descriptor: | CRISPR-associated helicase, Cas3 family, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQY
 
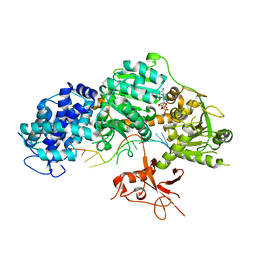 | | Crystal structure of T. fusca Cas3-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQX
 
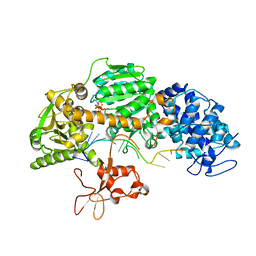 | | Crystal structure of T. fusca Cas3-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQW
 
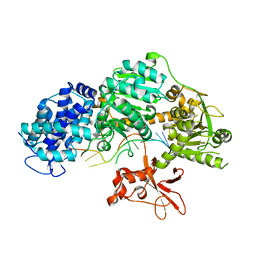 | | Crystal structure of T. fusca Cas3 | | Descriptor: | CRISPR-associated helicase, Cas3 family, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.664 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4Y1M
 
 | |
4Y1I
 
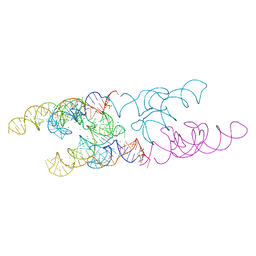 | | Lactococcus lactis yybP-ykoY Mn riboswitch bound to Mn2+ | | Descriptor: | BARIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Lactococcus lactis yybP-ykoY riboswitch, ... | | Authors: | Price, I.R, Ke, A. | | Deposit date: | 2015-02-07 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mn(2+)-Sensing Mechanisms of yybP-ykoY Orphan Riboswitches.
Mol.Cell, 57, 2015
|
|
4Y1J
 
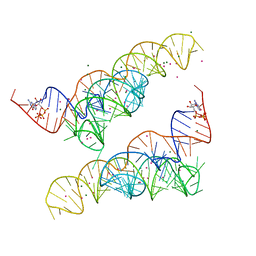 | |
3E5F
 
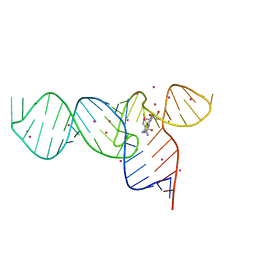 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with Se-SAM | | Descriptor: | SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3E5C
 
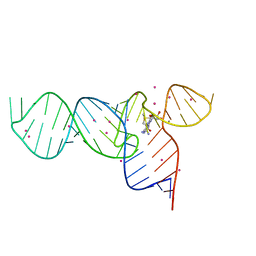 | | Crystal Structure of the SMK box (SAM-III) Riboswitch with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, SMK box (SAM-III) Riboswitch, STRONTIUM ION | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3E5E
 
 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4ES1
 
 | |
4F3M
 
 | | Crystal structure of CRISPR-associated protein | | Descriptor: | 1,2-ETHANEDIOL, BH0337 protein, SULFATE ION | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-05-09 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System.
Structure, 20, 2012
|
|
4KQY
 
 | | Bacillus subtilis yitJ S box/SAM-I riboswitch | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, YitJ S box/SAM-I riboswitch | | Authors: | Lu, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch
J.Mol.Biol., 404, 2010
|
|
6C27
 
 | | SAM-III riboswitch ON-state | | Descriptor: | COBALT HEXAMMINE(III), SAM-III riboswitch | | Authors: | Grigg, J.C, Price, I.R, Ke, A. | | Deposit date: | 2018-01-07 | | Release date: | 2019-01-09 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3.601 Å) | | Cite: | Evidence for two-tiered conformation selection in the SAM-III riboswitch
To Be Published
|
|
