7QJF
 
 | | Llp mutant C1G, lytic conversion lipoprotein of phage T5 | | Descriptor: | Lytic conversion lipoprotein | | Authors: | Degroux, S, Mestdach, E, Vives, C, Le Roy, A, Salmon, L, Herrman, T, Breyton, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Method: | SOLUTION NMR | | Cite: | Llp mutant C1G, lytic conversion lipoprotein of phage T5
To Be Published
|
|
8B14
 
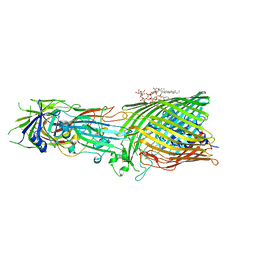 | | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, FhuA iron-ferrichrome transporter, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate, ... | | Authors: | Degroux, S, Effantin, G, Linares, R, Schoehn, G, Breyton, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger.
J.Virol., 97, 2023
|
|
8OKH
 
 | |
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
