7PFP
 
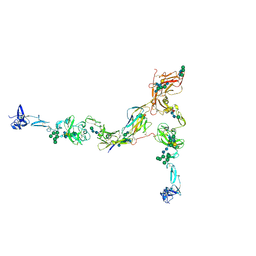 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5CSG
 
 | |
5HZV
 
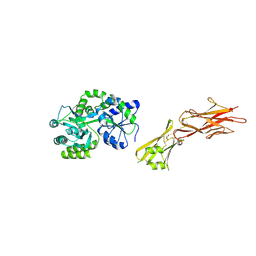 | | Crystal structure of the zona pellucida module of human endoglin/CD105 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Endoglin, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
3IRQ
 
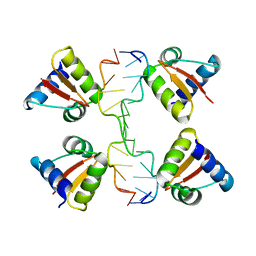 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IRR
 
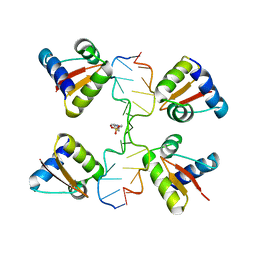 | | Crystal Structure of a Z-Z junction (with HEPES intercalating) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*A*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*G*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6FFT
 
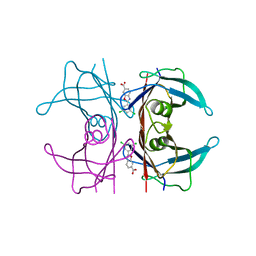 | | Neutron structure of human transthyretin (TTR) S52P mutant in complex with tafamidis at room temperature to 2A resolution (quasi-Laue) | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-02 | | Last modified: | 2019-03-20 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
5BUP
 
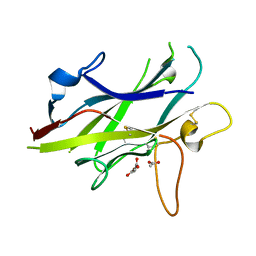 | | Crystal structure of the ZP-C domain of mouse ZP2 | | Descriptor: | ACETATE ION, Zona pellucida sperm-binding protein 2 | | Authors: | Nishimura, K, Jovine, L. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-27 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6QQK
 
 | |
6QQI
 
 | |
6QQJ
 
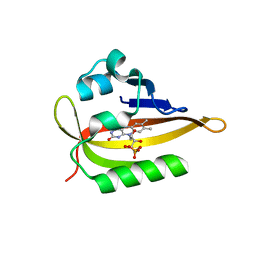 | |
6QSA
 
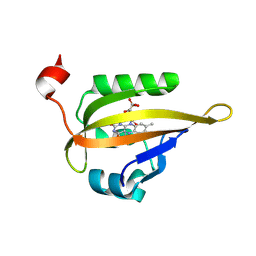 | |
5LTP
 
 | |
5LTQ
 
 | |
5LTR
 
 | |
7ZGY
 
 | | S-layer Deinoxanthin Binding Complex, C3 symmetry | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions.
J.Biol.Chem., 298, 2022
|
|
7ZGX
 
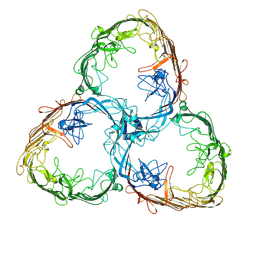 | | S-layer Deinoxanthin Binding Complex, C1 symmetry | | Descriptor: | S-layer protein SlpA | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions.
J.Biol.Chem., 298, 2022
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
8ACA
 
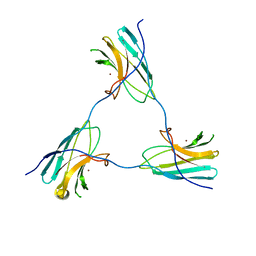 | | SDBC DR_0644 subunit, only-Cu Superoxide Dismutase | | Descriptor: | COPPER (II) ION, DR_0644, only-Cu Superoxide Dismutase | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8ACQ
 
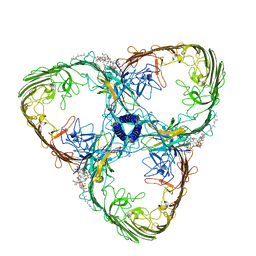 | | S-layer Deinoxanthin-Binding Complex (SDBC), subunit DR_2577 assembled with its SOD DR_0644 | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, DR_0644, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8AGD
 
 | | Full SDBC and SOD assembly | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Graca, A.T, Piano, D. | | Deposit date: | 2022-07-19 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
5NFE
 
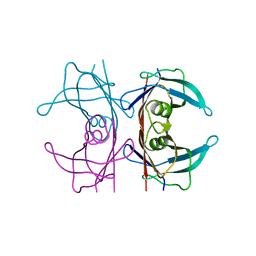 | | Neutron structure of human transthyretin (TTR) T119M mutant at room temperature to 1.85A resolution | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Ostermann, A, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-14 | | Release date: | 2019-01-02 | | Last modified: | 2019-03-20 | | Method: | NEUTRON DIFFRACTION (1.853 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
5NFW
 
 | | Neutron structure of human transthyretin (TTR) S52P mutant at room temperature to 1.8A resolution (quasi-Laue) | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2019-03-20 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
4X1T
 
 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | Authors: | Rocha, J, Breton, C. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
