2W37
 
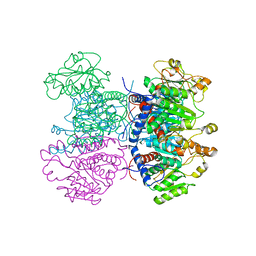 | | CRYSTAL STRUCTURE OF THE HEXAMERIC CATABOLIC ORNITHINE TRANSCARBAMYLASE FROM Lactobacillus hilgardii | | Descriptor: | NICKEL (II) ION, ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | de las Rivas, B, Fox, G.C, Angulo, I, Rodriguez, H, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hexameric Catabolic Ornithine Transcarbamylase from Lactobacillus Hilgardii: Structural Insights Into the Oligomeric Assembly and Metal Binding.
J.Mol.Biol., 393, 2009
|
|
4UEK
 
 | | Galactitol-1-phosphate 5-dehydrogenase from E. coli with Tris within the active site. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UEJ
 
 | | Closed state of galactitol-1-phosphate 5-dehydrogenase from E. coli in complex with glycerol. | | Descriptor: | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, GLYCEROL, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UEO
 
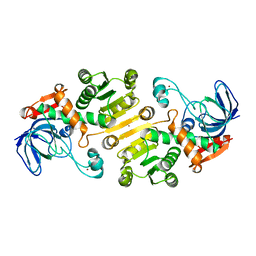 | | Open state of galactitol-1-phosphate 5-dehydrogenase from E. coli, with zinc in the catalytic site. | | Descriptor: | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2Y9G
 
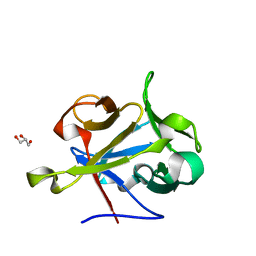 | | High-resolution Structural Insights on the Sugar-recognition and Fusion Tag Properties of a Versatile b-Trefoil Lectin Domain | | Descriptor: | GLYCEROL, HEMOLYTIC LECTIN LSLA, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Angulo, I, Acebron, I, de las Rivas, B, Munoz, R, Rodriguez, J.I, Menendez, M, Garcia, P, Tateno, H, Goldstein, I.J, Perez-Agote, B, Mancheno, J.M. | | Deposit date: | 2011-02-14 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-Resolution Structural Insights on the Sugar-Recognition and Fusion Tag Properties of a Versatile Beta-Trefoil Lectin Domain from the Mushroom Laetiporus Sulphureus.
Glycobiology, 21, 2011
|
|
2Y9F
 
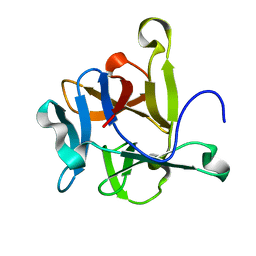 | | High-resolution Structural Insights on the Sugar-recognition and Fusion Tag Properties of a Versatile b-Trefoil Lectin Domain | | Descriptor: | HEMOLYTIC LECTIN LSLA | | Authors: | Angulo, I, Acebron, I, de las Rivas, B, Munoz, R, Rodriguez, J.I, Menendez, M, Garcia, P, Tateno, H, Goldstein, I.J, Perez-Agote, B, Mancheno, J.M. | | Deposit date: | 2011-02-14 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | High-Resolution Structural Insights on the Sugar-Recognition and Fusion Tag Properties of a Versatile Beta-Trefoil Lectin Domain from the Mushroom Laetiporus Sulphureus.
Glycobiology, 21, 2011
|
|
4A2C
 
 | | Crystal structure of galactitol-1-phosphate dehydrogenase from Escherichia coli | | Descriptor: | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, NICKEL (II) ION, ZINC ION | | Authors: | Alvarez, Y, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2011-09-26 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Crystal Structure of Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli K12 Provides Insights Into its Anomalous Behavior on Imac Processes
FEBS Lett., 586, 2012
|
|
2W22
 
 | | Activation Mechanism of Bacterial Thermoalkalophilic Lipases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CALCIUM ION, ... | | Authors: | Carrasco-Lopez, C, Godoy, C, De Las Rivas, B, Fernandez-Lorente, G, Palomo, J.M, Guisan, J.M, Fernandez-Lafuente, R, Hermoso, J.A. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements.
J. Biol. Chem., 284, 2009
|
|
2W2A
 
 | | Crystal Structure of p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2WSJ
 
 | | Crystal Structure of single point mutant Glu71Ser p-coumaric Acid Decarboxylase | | Descriptor: | BARIUM ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2009-09-07 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2W2F
 
 | | CRYSTAL STRUCTURE OF SINGLE POINT MUTANT ARG48GLN OF P-COUMARIC ACID DECARBOXYLASE FROM LACTOBACILLUS PLANTARUM STRUCTURAL INSIGHTS INTO THE ACTIVE SITE AND DECARBOXYLATION CATALYTIC MECHANISM | | Descriptor: | BARIUM ION, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-29 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
4C87
 
 | | Esterase LpEst1 from Lactobacillus plantarum WCFS1 | | Descriptor: | ESTERASE, GLYCEROL | | Authors: | Alvarez, Y, Esteban-Torres, M, Cortes-Cabrera, A, Gago, F, Acebron, I, Benavente, R, Mardo, K, de-las-Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2013-09-30 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Esterase Lpest1 from Lactobacillus Plantarum: A Novel and Atypical Member of the Alpha Beta Hydrolase Superfamily of Enzymes
Plos One, 9, 2014
|
|
4C89
 
 | | Crystal structure of carboxylesterase LpEst1 from Lactobacillus plantarum: high resolution model | | Descriptor: | ESTERASE, GLYCEROL, MALONATE ION | | Authors: | Alvarez, Y, Esteban-Torres, M, Cortes-Cabrera, A, Gago, F, Acebron, I, Benavente, R, Mardo, K, de-las-Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2013-09-30 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Esterase Lpest1 from Lactobacillus Plantarum: A Novel and Atypical Member of the Alpha Beta Hydrolase Superfamily of Enzymes
Plos One, 9, 2014
|
|
4C88
 
 | | Esterase LpEst1 from Lactobacillus plantarum: native structure | | Descriptor: | ESTERASE | | Authors: | Alvarez, Y, Esteban-Torres, M, Cortes-Cabrera, A, Gago, F, Acebron, I, Benavente, R, Mardo, K, de-las-Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2013-09-30 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Esterase Lpest1 from Lactobacillus Plantarum: A Novel and Atypical Member of the Alpha Beta Hydrolase Superfamily of Enzymes
Plos One, 9, 2014
|
|
4BZZ
 
 | | Complete crystal structure of carboxylesterase Cest-2923 from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, LIPASE/ESTERASE, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
4C01
 
 | | Complete crystal structure of carboxylesterase Cest-2923 (lp_2923) from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, CEST-2923, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
5NAV
 
 | |
6YQ4
 
 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
5NAQ
 
 | |
2W2B
 
 | | Crystal Structure of single point mutant Tyr20Phe p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | ACETATE ION, ISOPROPYL ALCOHOL, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
4BZW
 
 | | Complete crystal structure of the carboxylesterase Cest-2923 (lp_2923) from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, LIPASE/ESTERASE, SULFATE ION, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, delasRivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
3NAD
 
 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
