8A6J
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8ACH
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8B9U
 
 | |
8B9O
 
 | |
2FFH
 
 | | THE SIGNAL SEQUENCE BINDING PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | CADMIUM ION, PROTEIN (FFH), SULFATE ION | | Authors: | Keenan, R.J, Freymann, D.M, Walter, P, Stroud, R.M. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the signal sequence binding subunit of the signal recognition particle.
Cell(Cambridge,Mass.), 94, 1998
|
|
5KUT
 
 | | hMiro2 C-terminal GTPase domain, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSY
 
 | | hMiro1 C-domain GDP Complex P41212 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
6D71
 
 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | Authors: | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2018-04-23 | | Release date: | 2019-10-09 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.7180779 Å) | | Cite: | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
5KSZ
 
 | | hMiro EF hand and cGTPase domains in the GMPPCP-bound state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mitochondrial Rho GTPase 1, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KTY
 
 | | hMiro EF hand and cGTPase domains, GDP and Ca2+ bound state | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
4LDD
 
 | |
4LDM
 
 | |
5L40
 
 | | polyketide ketoreductase SimC7 - apo crystal form 1 | | Descriptor: | polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5KSP
 
 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
4LDI
 
 | |
4LDB
 
 | |
5KP9
 
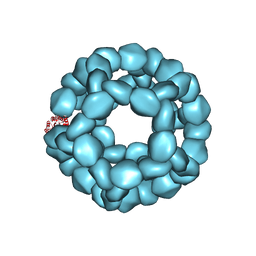 | | Structure of Nanoparticle Released from Enveloped Protein Nanoparticle | | Descriptor: | EPN-01* | | Authors: | Votteler, J, Ogohara, C, Yi, S, Hsia, Y, Natterman, U, Belnap, D.M, King, N.P, Sundquist, W.I. | | Deposit date: | 2016-07-02 | | Release date: | 2016-12-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Designed proteins induce the formation of nanocage-containing extracellular vesicles.
Nature, 540, 2016
|
|
4LD8
 
 | |
5L4L
 
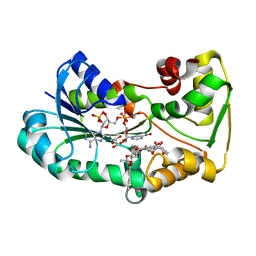 | | polyketide ketoreductase SimC7 - ternary complex with NADP+ and 7-oxo-SD8 | | Descriptor: | 7-oxo-simocyclinone D8, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5KSO
 
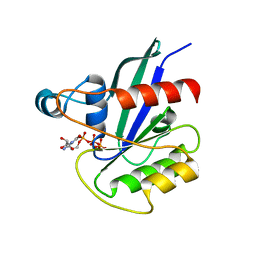 | | hMiro1 C-domain GDP-Pi Complex P3121 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1, PHOSPHATE ION | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KU1
 
 | | hMiro1 EF hand and cGTPase domains in the GDP-bound state | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5L3Z
 
 | | polyketide ketoreductase SimC7 - binary complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5L45
 
 | | polyketide ketoreductase SimC7 - apo crystal form 2 | | Descriptor: | GLYCEROL, SimC7, TETRAETHYLENE GLYCOL | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
5LGV
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5LQD
 
 | | Trehalose-6-phosphate synthase, GDP-glucose-dependent OtsA | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase | | Authors: | Miah, F, Asencion Diez, M.D, Stevenson, C.E.M, Lawson, D.M, Iglesias, A.A, Bornemann, S. | | Deposit date: | 2016-08-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Production and Utilization of GDP-glucose in the Biosynthesis of Trehalose 6-Phosphate by Streptomyces venezuelae.
J. Biol. Chem., 292, 2017
|
|
