3TTF
 
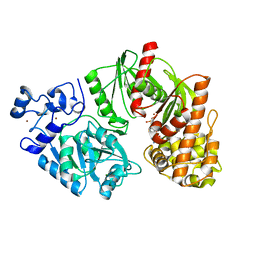 | | Crystal structure of E. coli HypF with AMP and carbamoyl phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TSQ
 
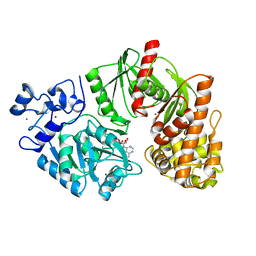 | | Crystal structure of E. coli HypF with ATP and Carbamoyl phosphate | | Descriptor: | 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
1JHN
 
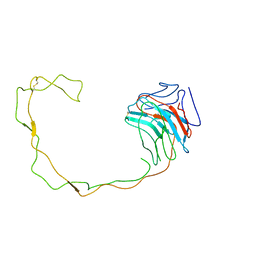 | | Crystal Structure of the Lumenal Domain of Calnexin | | Descriptor: | CALCIUM ION, calnexin | | Authors: | Schrag, J.D, Bergeron, J.M, Li, Y, Borisova, S, Hahn, M, Thomas, D.Y, Cygler, M. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of calnexin, an ER chaperone involved in quality control of protein folding.
Mol.Cell, 8, 2001
|
|
3TSP
 
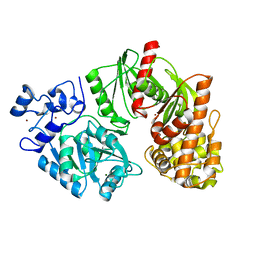 | | Crystal structure of E. coli HypF | | Descriptor: | MAGNESIUM ION, Transcriptional regulatory protein, ZINC ION | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TSU
 
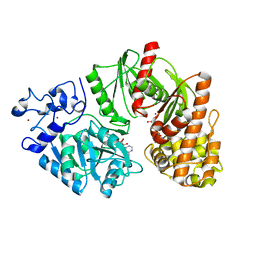 | | Crystal structure of E. coli HypF with AMP-PNP and carbamoyl phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TTC
 
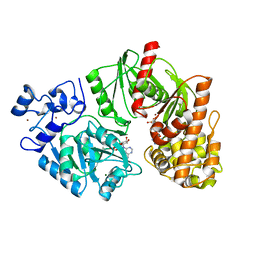 | | Crystal structure of E. coli HypF with ADP and carbamoyl phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TTD
 
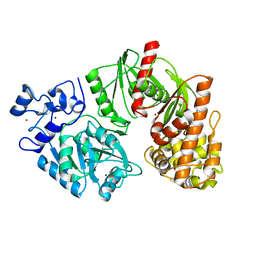 | | Crystal structure of E. coli HypF with AMP-CPP and carbamoyl phosphate | | Descriptor: | MAGNESIUM ION, Transcriptional regulatory protein, ZINC ION | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
2OVB
 
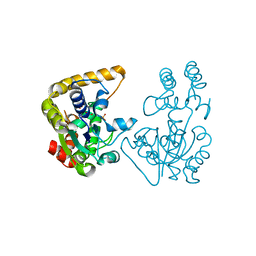 | | Crystal Structure of StaL-sulfate complex | | Descriptor: | SULFATE ION, StaL | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of StaL, a glycopeptide antibiotic sulfotransferase from Streptomyces toyocaensis.
J.Biol.Chem., 282, 2007
|
|
2OV8
 
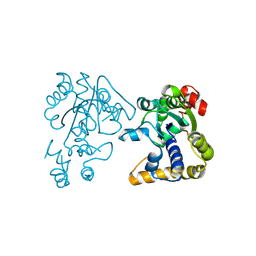 | |
2OVF
 
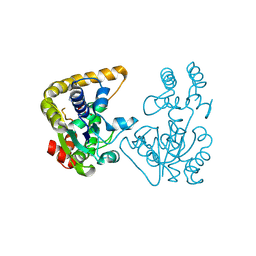 | | Crystal Structure of StaL-PAP complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, StaL | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of StaL, a glycopeptide antibiotic sulfotransferase from Streptomyces toyocaensis.
J.Biol.Chem., 282, 2007
|
|
2Q1F
 
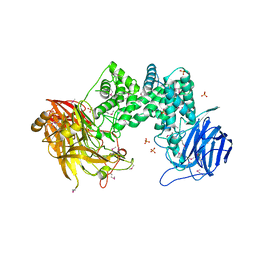 | |
2RB9
 
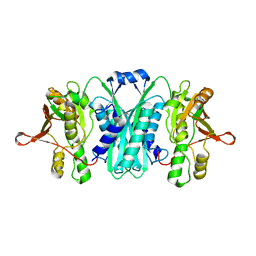 | | Crystal structure of E.coli HypE | | Descriptor: | HypE protein | | Authors: | Asinas, A.E, Rangarajan, E.S, Min, T, Matte, A, Proteau, A, Munger, C, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
3LVJ
 
 | | Crystal Structure of E.coli IscS-TusA complex (form 1) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
1YQC
 
 | | Crystal Structure of Ureidoglycolate Hydrolase (AllA) from Escherichia coli O157:H7 | | Descriptor: | GLYOXYLIC ACID, Ureidoglycolate hydrolase | | Authors: | Raymond, S, Tocilj, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase (AllA) from Escherichia coli O157:H7
Proteins, 61, 2005
|
|
1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNH
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
3LVK
 
 | | Crystal Structure of E.coli IscS-TusA complex (form 2) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVM
 
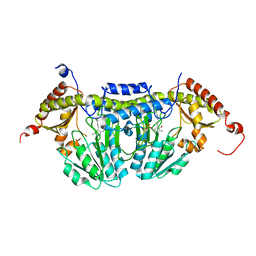 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
1ZPS
 
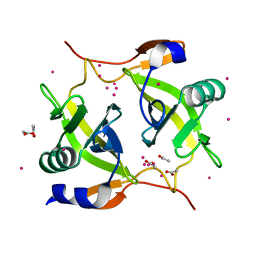 | | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | | Descriptor: | ACETIC ACID, CADMIUM ION, Phosphoribosyl-AMP cyclohydrolase | | Authors: | Sivaraman, J, Myers, R.S, Boju, L, Sulea, T, Cygler, M, Davisson, V.J, Schrag, J.D, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Methanobacterium thermoautotrophicum Phosphoribosyl-AMP Cyclohydrolase HisI.
Biochemistry, 44, 2005
|
|
2AHV
 
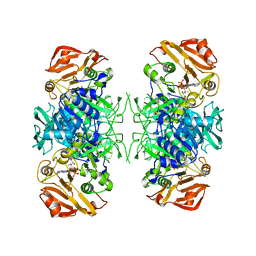 | | Crystal Structure of Acyl-CoA transferase from E. coli O157:H7 (YdiF)-thioester complex with CoA- 1 | | Descriptor: | COENZYME A, putative enzyme YdiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
3IKW
 
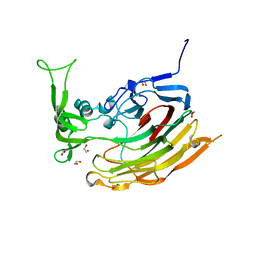 | | Structure of Heparinase I from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heparin lyase I | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
2AHU
 
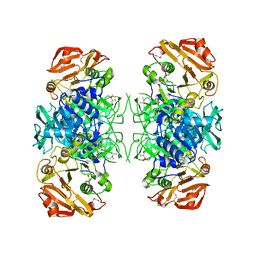 | | Crystal structure of Acyl-CoA transferase (YdiF) apoenzyme from Escherichia coli O157:H7. | | Descriptor: | putative enzyme ydiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
1K94
 
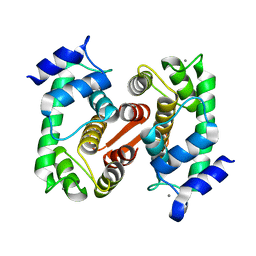 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K95
 
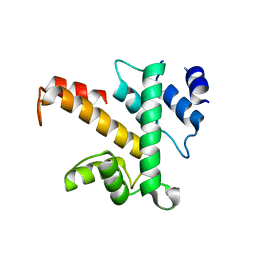 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3PT5
 
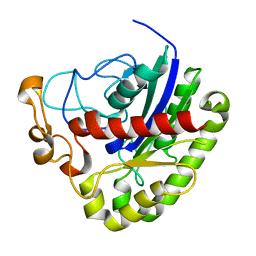 | | Crystal structure of NanS | | Descriptor: | NANS (YJHS), A 9-O-acetyl N-acetylneuraminic acid esterase | | Authors: | Ruane, K.M, Rangarajan, E.S, Proteau, A, Schrag, J.D, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-12-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7.
Protein Sci., 20, 2011
|
|
