2FNU
 
 | | PseC aminotransferase with external aldimine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, aminotransferase | | Authors: | Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FN6
 
 | | Helicobacter pylori PseC, aminotransferase involved in the biosynthesis of pseudoaminic acid | | Descriptor: | AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Cygler, M, Lunin, V.V, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
1CB8
 
 | | CHONDROITINASE AC LYASE FROM FLAVOBACTERIUM HEPARINUM | | Descriptor: | CALCIUM ION, GLYCEROL, PROTEIN (CHONDROITINASE AC), ... | | Authors: | Fethiere, J, Eggimann, B, Cygler, M. | | Deposit date: | 1999-03-02 | | Release date: | 1999-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of chondroitin AC lyase, a representative of a family of glycosaminoglycan degrading enzymes.
J.Mol.Biol., 288, 1999
|
|
4WL1
 
 | |
8EW1
 
 | | Structure of Bacple_01703-E145L | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
8SWI
 
 | |
8EP4
 
 | | Structure of Bacple_01703 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
8SR6
 
 | |
1A4I
 
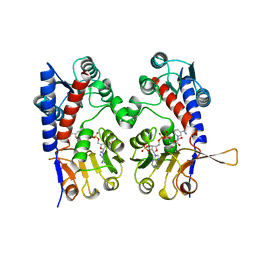 | | HUMAN TETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE / METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Allaire, M, Li, Y, Mackenzie, R.E, Cygler, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 3-D structure of a folate-dependent dehydrogenase/cyclohydrolase bifunctional enzyme at 1.5 A resolution.
Structure, 6, 1998
|
|
7PCK
 
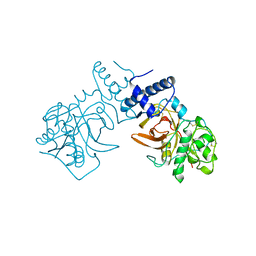 | |
7SNJ
 
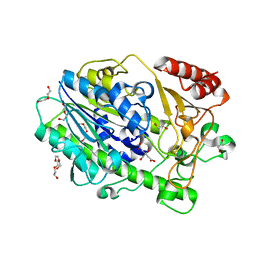 | | Structure of Bacple_01701, a 6-O-galactose porphyran sulfatase | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Arylsulfatase, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
7SNK
 
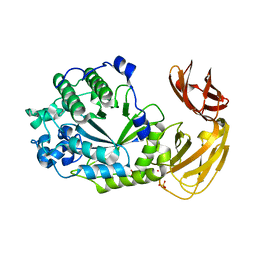 | | Structure of Bacple_01702, a GH29 family glycoside hydrolase | | Descriptor: | Alpha-L-fucosidase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
7SNO
 
 | | Structure of Bacple_01701(H214N), a 6-O-galactose porphyran sulfatase | | Descriptor: | 1,2-ETHANEDIOL, 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, Arylsulfatase, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
6P0X
 
 | |
7RTK
 
 | | Structure of the (NIAU)2 complex with N-terminal mutation of ISCU2 Y35D at 2.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2021-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
1AC5
 
 | | CRYSTAL STRUCTURE OF KEX1(DELTA)P, A PROHORMONE-PROCESSING CARBOXYPEPTIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, KEX1(DELTA)P | | Authors: | Shilton, B.H, Thomas, D.Y, Cygler, M. | | Deposit date: | 1997-02-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Kex1deltap, a prohormone-processing carboxypeptidase from Saccharomyces cerevisiae.
Biochemistry, 36, 1997
|
|
4FZW
 
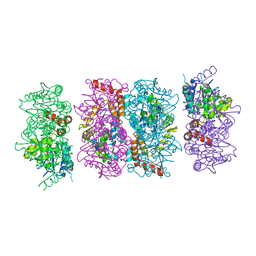 | | Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex from E.coli | | Descriptor: | 1,2-epoxyphenylacetyl-CoA isomerase, 2,3-dehydroadipyl-CoA hydratase, GLYCEROL | | Authors: | Grishin, A.M, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-07-08 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein-Protein Interactions in the beta-Oxidation Part of the Phenylacetate
Utilization Pathway. Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex
J.Biol.Chem., 287, 2012
|
|
7TZK
 
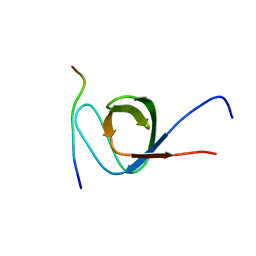 | | EPS8 SH3 Domain with NleH1 PxxDY Motif | | Descriptor: | Epidermal growth factor receptor kinase substrate 8, T3SS secreted effector NleH homolog | | Authors: | Grishin, A.M, Cygler, M. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UPS
 
 | |
7LSV
 
 | |
2ZL1
 
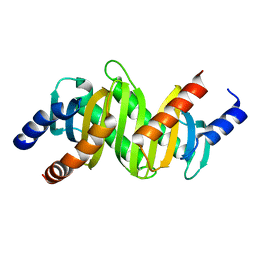 | | MP1-p14 Scaffolding complex | | Descriptor: | Mitogen-activated protein kinase kinase 1-interacting protein 1, Mitogen-activated protein-binding protein-interacting protein | | Authors: | Schrag, J.D, Cygler, M, Munger, C, Magloire, A. | | Deposit date: | 2008-04-02 | | Release date: | 2008-06-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular dynamics-solvated interaction energy studies of protein-protein interactions: the MP1-p14 scaffolding complex.
J.Mol.Biol., 379, 2008
|
|
5KDG
 
 | | Crystal Structure of Salmonella Typhimurium Effector GtgE | | Descriptor: | GLYCEROL, Gifsy-2 prophage protein, SULFATE ION | | Authors: | Kozlov, G, Xu, C, Wong, K, Gehring, K, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the Salmonella Typhimurium Effector GtgE.
PLoS ONE, 11, 2016
|
|
5JMF
 
 | | Heparinase III-BT4657 gene product | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Heparinase III protein, ... | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
5JMD
 
 | | Heparinase III-BT4657 gene product, Methylated Lysines | | Descriptor: | Heparinase III protein, MAGNESIUM ION | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
2VHE
 
 | | PglD-CoA complex: An acetyl transferase from Campylobacter jejuni | | Descriptor: | ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Rangarajan, E.S, Ruane, K.M, Sulea, T, Watson, D.C, Proteau, A, Leclerc, S, Cygler, M, Matte, A, Young, N.M. | | Deposit date: | 2007-11-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Active Site Residues of Pgld, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter Jejuni
Biochemistry, 47, 2008
|
|
