1AWO
 
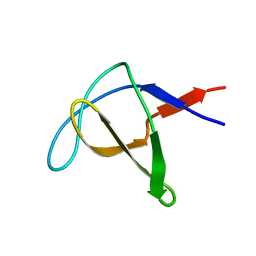 | |
1Z66
 
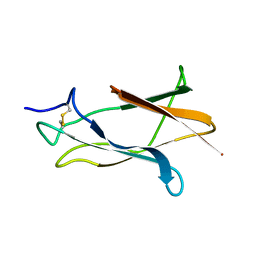 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | Descriptor: | Major envelope protein E | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
7SGT
 
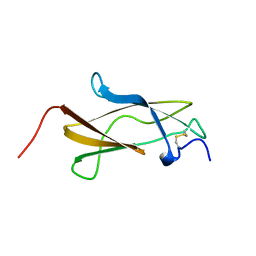 | |
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1JEG
 
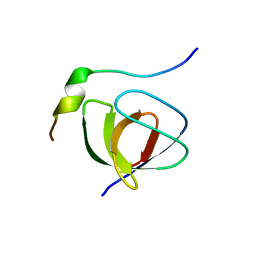 | | Solution structure of the SH3 domain from C-terminal Src Kinase complexed with a peptide from the tyrosine phosphatase PEP | | Descriptor: | HEMATOPOIETIC CELL PROTEIN-TYROSINE PHOSPHATASE 70Z-PEP, TYROSINE-PROTEIN KINASE CSK | | Authors: | Ghose, R, Shekhtman, A, Goger, M.J, Ji, H, Cowburn, D. | | Deposit date: | 2001-06-17 | | Release date: | 2001-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel, specific interaction involving the Csk SH3 domain and its natural ligand.
Nat.Struct.Biol., 8, 2001
|
|
2K6X
 
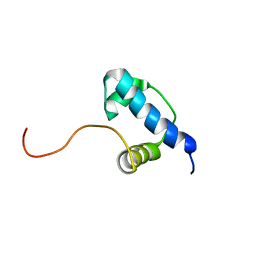 | | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1- Induced Compacted Structure | | Descriptor: | RNA polymerase sigma factor rpoD | | Authors: | Schwartz, E.C, Shekhtman, A, Dutta, K, Pratt, M.R, Cowburn, D, Darst, S, Muir, T.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1 - Induced Compacted Structure
Chem.Biol., 15, 2008
|
|
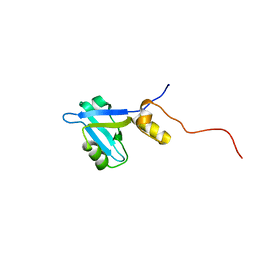
2KJD
 
 | |
6AX5
 
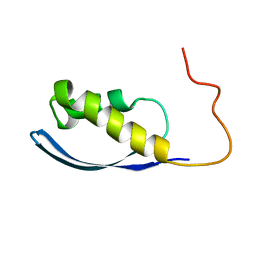 | | RPT1 region of INI1/SNF5/SMARCB1_HUMAN - SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Girvin, M.E, Cahill, S.M, Harris, R, Cowburn, D, Spira, M, Wu, X, Prakash, R, Bernowitz, M, Almo, S.C, Kalpana, G.V. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.
Nat Commun, 12, 2021
|
|
6DSL
 
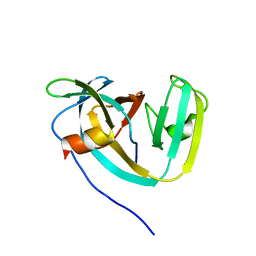 | | Consensus engineered intein (Cat) with atypical split site | | Descriptor: | Consensus engineered intein CatC, Consensus engineered intein CatN | | Authors: | Sekar, G, Stevens, A.J, Muir, T.W, Cowburn, D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An Atypical Mechanism of Split Intein Molecular Recognition and Folding.
J. Am. Chem. Soc., 140, 2018
|
|
4HDI
 
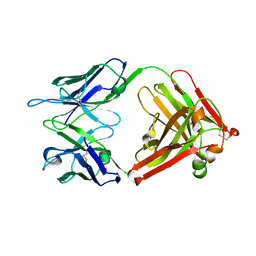 | | Crystal Structure of 3E5 IgG3 FAB from mus musculus | | Descriptor: | Ig heavy chain V region RF, Ig gamma-3 chain C region, Kappa light chain variable region, ... | | Authors: | Janda, A, Eryilmaz, E, Kim, J, Cordero, R.J.B, Cowburn, D, Casadevall, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Global structures of IgG isotypes expressing identical variable regions.
Mol.Immunol., 56, 2013
|
|
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
3EAZ
 
 | |
3EAC
 
 | |
1AB2
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY 2 DOMAIN OF C-ABL | | Descriptor: | C-ABL TYROSINE KINASE SH2 DOMAIN | | Authors: | Overduin, M, Rios, C.B, Mayer, B.J, Baltimore, D, Cowburn, D. | | Deposit date: | 1993-07-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the src homology 2 domain of c-abl.
Cell(Cambridge,Mass.), 70, 1992
|
|
2GGR
 
 | |
2GG1
 
 | | NMR solution structure of domain III of the E-protein of tick-borne Langat flavivirus (includes RDC restraints) | | Descriptor: | Genome polyprotein | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
3BRH
 
 | |
2L6E
 
 | | NMR Structure of the monomeric mutant C-terminal domain of HIV-1 Capsid in complex with stapled peptide Inhibitor | | Descriptor: | Capsid protein p24, NYAD-13 stapled peptide inhibitor | | Authors: | Bhattacharya, S, Zhang, H, Debnath, A.K, Cowburn, D. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid.
J.Biol.Chem., 283, 2008
|
|
1DDB
 
 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (BID) | | Authors: | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | Deposit date: | 1999-02-19 | | Release date: | 1999-08-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
2M0U
 
 | | Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0V
 
 | | Complex structure of C-terminal CFTR peptide and extended PDZ2 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0T
 
 | |
2KRG
 
 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
1BAK
 
 | |
1A6S
 
 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
