1NEI
 
 | | Solution NMR Structure of Protein yoaG from Escherichia coli. Ontario Centre for Structural Proteomics Target EC0264_1_60; Northeast Structural Genomics Consortium Target ET94. | | Descriptor: | hypothetical protein yoaG | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein dimer encoded by the Yoag gene from Escherichia coli
To be published
|
|
2JRA
 
 | | A novel domain-swapped solution NMR structure of protein RPA2121 from Rhodopseudomonas palustris. Northeast Structural Genomics Target RpT6 | | Descriptor: | Protein RPA2121 | | Authors: | Wu, B, Yee, A, Lemak, A, Cort, J, Bansal, S, Semest, A, Guido, V, Kennedy, M.A, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-21 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A novel domain-swapped solution NMR structure of protein RPA2121 from Rhodopseudomonas palustris.
To be Published
|
|
2K0Z
 
 | | Solution NMR structure of protein hp1203 from Helicobacter pylori 26695. Northeast Structural Genomics Consortium (NESG) target PT1/Ontario Center for Structural Proteomics target hp1203 | | Descriptor: | Uncharacterized protein hp1203 | | Authors: | Wu, B, Yee, A, Lemak, A, Cort, J, Semest, A, Kenney, M.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein hp1203 from Helicobacter pylori 26695. Northeast Structural Genomics Consortium (NESG) target PT1/Ontario Center for Structural Proteomics target hp1203.
To be Published
|
|
1AO1
 
 | | INTERACTIONS OF DEGLYCOSYLATED COBALT(III)-PEPLEOMYCIN WITH DNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | COBALT (III)-DEGLYCOPEPLEOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | Deposit date: | 1997-07-16 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of deglycosylated cobalt(III)-pepleomycin (green form) with DNA based on NMR structural studies,.
Biochemistry, 36, 1997
|
|
1AO2
 
 | | cobalt(III)-deglycopepleomycin determined by NMR studies | | Descriptor: | AGLYCON OF PEPLOMYCIN, COBALT (III) ION, HYDROGEN PEROXIDE | | Authors: | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | Deposit date: | 1997-07-16 | | Release date: | 1999-07-30 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
1AO4
 
 | | COBALT(III)-PEPLOMYCIN COMPLEX DETERMINED BY NMR STUDIES | | Descriptor: | 3-O-carbamoyl-alpha-D-mannopyranose-(1-2)-alpha-L-gulopyranose, AGLYCON OF PEPLOMYCIN, COBALT (III) ION, ... | | Authors: | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | Deposit date: | 1997-07-16 | | Release date: | 1999-07-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
8FCI
 
 | | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lavallee, T, Mehta, K, Young, A, Cortes, J, Isaacson, B, Brylewski, J, Schlegel, F, Doti, L, Battaile, K, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine
To Be Published
|
|
8FCH
 
 | | Apo Structure of (N1G37) Methyltransferase from Mycobacterium avium | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Isaacson, B, Brylewski, J, Schlegel, F, Cortes, J, Lavallee, T, Mehta, K, Young, A, Doti, L, Battaile, K.P, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
To Be Published
|
|
2MTW
 
 | | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit into HLA-DR 1*03 molecules induces antibody production and protection | | Descriptor: | Erythrocyte-binding antigen 175 | | Authors: | Cifuentes, G, Salazar, L, Vargas, L, Parra, C, Vanegas, M, Cortes, J, Sandoval, M, Patarroyo, M.E. | | Deposit date: | 2014-09-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit HLA-DR 1*03 molecules induces antibody production and protection
To be Published
|
|
3IL5
 
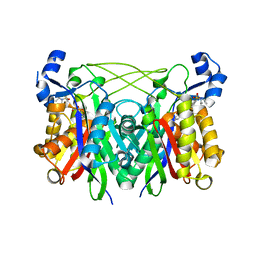 | | Structure of E. faecalis FabH in complex with 2-({4-bromo-3-[(diethylamino)sulfonyl]benzoyl}amino)benzoic acid | | Descriptor: | 2-({[4-bromo-3-(diethylsulfamoyl)phenyl]carbonyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL6
 
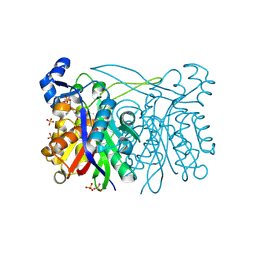 | | Structure of E. faecalis FabH in complex with 2-({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxybenzoyl}amino)benzoic acid | | Descriptor: | 2-[({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxyphenyl}carbonyl)amino]benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SULFATE ION | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL9
 
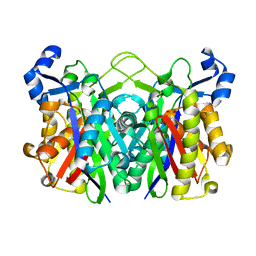 | | Structure of E. coli FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL7
 
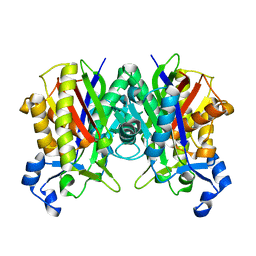 | | Crystal structure of S. aureus FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL3
 
 | | Structure of Haemophilus influenzae FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL4
 
 | | Structure of E. faecalis FabH in complex with acetyl CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, ACETYL COENZYME *A | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
2L0C
 
 | | Solution NMR Structure of protein STY4237 (residues 36-120) from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR115 | | Descriptor: | Putative membrane protein | | Authors: | Cort, J.R, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein STY4237 (residues 36-120) from
Salmonella enterica, Northeast Structural Genomics Consortium Target SlR115
To be Published
|
|
2L0D
 
 | | Solution NMR Structure of putative cell surface protein MA_4588 (272-376 domain) from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR254A | | Descriptor: | Cell surface protein | | Authors: | Cort, J.R, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of putative cell surface protein MA_4588
(272-376 domain) from Methanosarcina acetivorans, Northeast Structural
Genomics Consortium Target MvR254A
To be Published
|
|
8SCE
 
 | |
4NY9
 
 | | Crystal Structure Of the Human PXR-LBD In Complex With N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-3-hydroxy-3-methylbutanamide | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-3-hydroxy-3-methylbutanamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of the CCR1 antagonist, BMS-817399, for the treatment of rheumatoid arthritis.
J.Med.Chem., 57, 2014
|
|
8SCW
 
 | |
8SCV
 
 | |
6YQ4
 
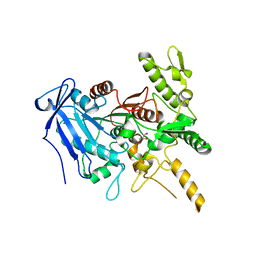 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
