6GIQ
 
 | | Saccharomyces cerevisiae respiratory supercomplex III2IV | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Rathore, S, Berndtsson, J, Conrad, J, Ott, M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the yeast respiratory supercomplex.
Nat. Struct. Mol. Biol., 26, 2019
|
|
7BOB
 
 | | Exo-beta-1,4-mannosidase Op5Man5 from Opitutaceae bacterium strain TAV5 | | Descriptor: | Endo-beta-mannanase | | Authors: | Kalyani, D.C, Reichenbach, T, Keskitalo, M.M, Conrad, J, Aspeborg, H, Divne, C. | | Deposit date: | 2021-01-24 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a homotrimeric verrucomicrobial exo - beta -1,4-mannosidase active in the hindgut of the wood-feeding termite Reticulitermes flavipes .
J Struct Biol X, 5, 2021
|
|
5N0C
 
 | | Crystal structure of the tetanus neurotoxin in complex with GM1a | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tetanus toxin, ZINC ION, ... | | Authors: | Masuyer, G, Conrad, J, Stenmark, P. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the tetanus toxin reveals pH-mediated domain dynamics.
EMBO Rep., 18, 2017
|
|
5N0B
 
 | | Crystal structure of the tetanus neurotoxin in complex with GD1a | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin, ... | | Authors: | Masuyer, G, Conrad, J, Stenmark, P. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the tetanus toxin reveals pH-mediated domain dynamics.
EMBO Rep., 18, 2017
|
|
5M1S
 
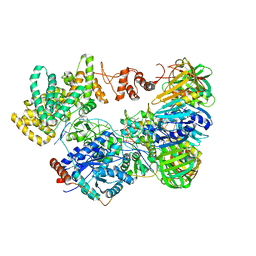 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5FKV
 
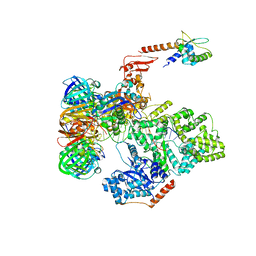 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKW
 
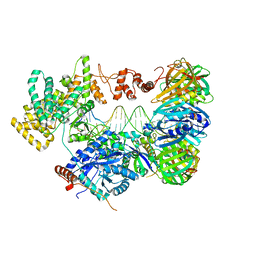 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKU
 
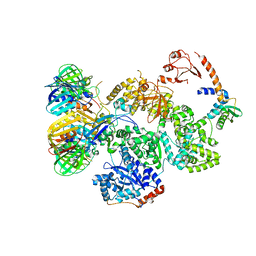 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
7Z1H
 
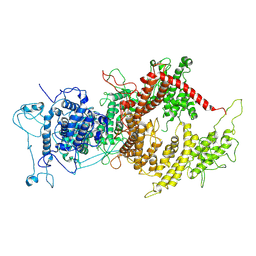 | | VAR2CSA APO | | Descriptor: | VAR2CSA APO | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
7Z12
 
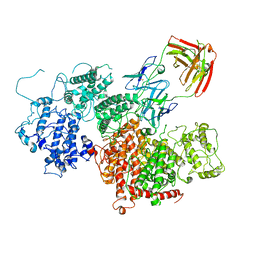 | | VAR2 complex with PAM1.4 | | Descriptor: | PAM1.4, Heavy Chain, light Chain, ... | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
6ZU6
 
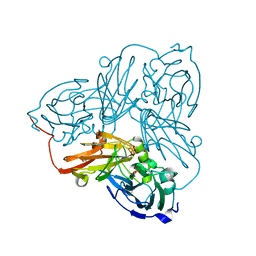 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUB
 
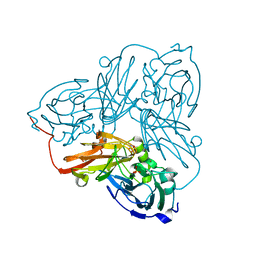 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 2 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUT
 
 | | Cu nitrite reductase MSOX series at 170K, dose point 5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Anyonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUA
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 4 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUD
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 3 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
3ZCJ
 
 | |
3ZCI
 
 | |
4CII
 
 | | Crystal structure of N-terminally truncated Helicobacter pylori T4SS Protein CagL as domain swapped dimer | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CAG PATHOGENICITY ISLAND PROTEIN (CAG18) | | Authors: | Barden, S, Schomburg, B, Niemann, H.H. | | Deposit date: | 2013-12-09 | | Release date: | 2014-02-19 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a Three-Dimensional Domain-Swapped Dimer of the Helicobacter Pylori Type Iv Secretion System Pilus Protein Cagl.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
