2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y2A
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | Descriptor: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5FVG
 
 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
2Y3L
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 2 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5FOZ
 
 | | De novo structure of the binary mosquito larvicide BinAB at pH 10 | | Descriptor: | LARVICIDAL TOXIN PROTEIN, SODIUM ION, TOXIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5G37
 
 | | MR structure of the binary mosquito larvicide BinAB at pH 5 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2016-04-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5FOY
 
 | | De novo structure of the binary mosquito larvicide BinAB at pH 7 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5G1K
 
 | | A triple mutant of DsbG engineered for denitrosylation | | Descriptor: | SULFATE ION, THIOL DISULFIDE INTERCHANGE PROTEIN DSBG | | Authors: | Tamu Dufe, V, Van Molle, I, Lafaye, C, Wahni, K, Boudier, A, Leroy, P, Collet, J.F, Messens, J. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Sulfur Denitrosylation by an Engineered Trx-Like Dsbg Enzyme Identifies Nucleophilic Cysteine Hydrogen Bonds as Key Functional Determinant.
J.Biol.Chem., 291, 2016
|
|
5G1L
 
 | | A double mutant of DsbG engineered for denitrosylation | | Descriptor: | SULFATE ION, THIOL DISULFIDE INTERCHANGE PROTEIN DSBG | | Authors: | Tamu Dufe, V, Van Molle, I, Lafaye, C, Wahni, K, Boudier, A, Leroy, P, Collet, J.F, Messens, J. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfur Denitrosylation by an Engineered Trx-Like Dsbg Enzyme Identifies Nucleophilic Cysteine Hydrogen Bonds as Key Functional Determinant.
J.Biol.Chem., 291, 2016
|
|
2KZ8
 
 | | Solution NMR structure of MqsA, a protein from E. coli, containing a Zinc finger, N-terminal and a Helix Turn-Helix C-terminal domain | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygiT | | Authors: | Papadopoulos, E, Vlamis-Gardikas, A, Graslund, A, Billeter, M, Holmgren, A, Collet, J. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biophysical properties of MqsA, a Zn-containing antitoxin from Escherichia coli
Biochim.Biophys.Acta, 2012
|
|
2J4C
 
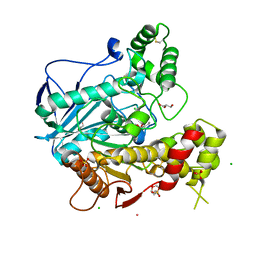 | | Structure of human Butyrylcholinesterase in complex with 10mM HgCl2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Colletier, J.P, Frasco, M.F, Carvalho, F, Guilhermino, L, Stojan, J, Fournier, D, Weik, M. | | Deposit date: | 2006-08-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanisms of Cholinesterase Inhibition by Inorganic Mercury.
FEBS J., 274, 2007
|
|
4D65
 
 | |
4D64
 
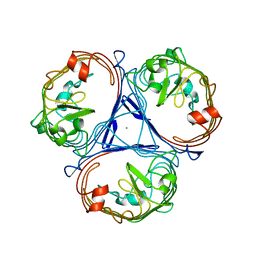 | |
4C69
 
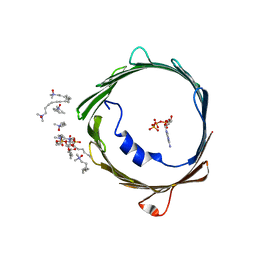 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4A16
 
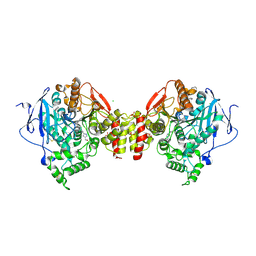 | | Structure of mouse Acetylcholinesterase complex with Huprine derivative | | Descriptor: | (1-{4-[(7S,11S)-12-AMINO-3-CHLORO-6,7,10,11-TETRAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-9-YL]BUTYL}-1H-1,2,3-TRIAZOL-4-YL)METHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Carletti, E, Colletier, J.P, Nachon, F, Weik, M, Ronco, C, Jean, L, Renard, P.Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Huprine Derivatives as Sub-Nanomolar Human Acetylcholinesterase Inhibitors: From Rational Design to Validation by X-Ray Crystallography.
Chemmedchem, 7, 2012
|
|
6H5Q
 
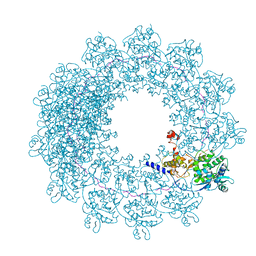 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4BGV
 
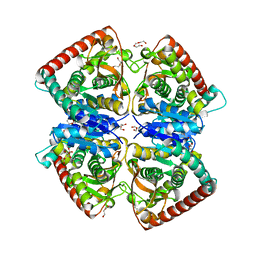 | |
4BGU
 
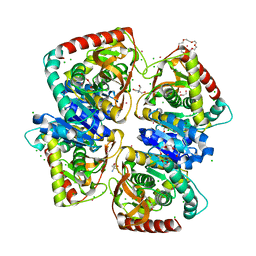 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
4CL3
 
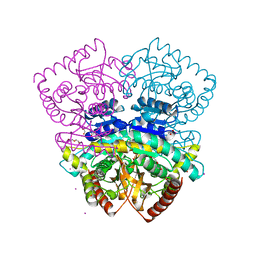 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
