1YXY
 
 | | Crystal Structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes (APC29713) Structural genomics, MCSG | | Descriptor: | Putative N-acetylmannosamine-6-phosphate 2-epimerase | | Authors: | Rotella, F.J, Zhang, R.G, Lezondra, L.E.O, Collart, F.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes
To be Published
|
|
1YSJ
 
 | | Crystal Structure of Bacillus Subtilis YXEP Protein (APC1829), a Dinuclear Metal Binding Peptidase from M20 Family | | Descriptor: | NICKEL (II) ION, protein yxeP | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Bacillus Subtilis YXEP Protein, a Dinuclear Metal Binding Peptidase from M20 Family
To be Published
|
|
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
1ZKP
 
 | | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease | | Descriptor: | CHLORIDE ION, SODIUM ION, ZINC ION, ... | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family
Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease
To be Published
|
|
1Z05
 
 | | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein. | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, SULFATE ION, ... | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-08 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein.
To be Published
|
|
1Y2I
 
 | | Crystal Structure of MCSG Target APC27401 from Shigella flexneri | | Descriptor: | Hypothetical Protein S0862 | | Authors: | Brunzelle, J.S, Minasov, G, Yang, X, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-22 | | Release date: | 2005-01-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC27401 from Shigella flexneri
To be Published
|
|
1ZSW
 
 | | Crystal Structure of Bacillus cereus Metallo Protein from Glyoxalase family | | Descriptor: | BETA-MERCAPTOETHANOL, Glyoxalase family protein, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Bacillus cereus Metallo Protein from Glyoxalase family
To be Published
|
|
3TX6
 
 | | The Structure of a putative ABC-transporter periplasmic component from Rhodopseudomonas palustris | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4EYQ
 
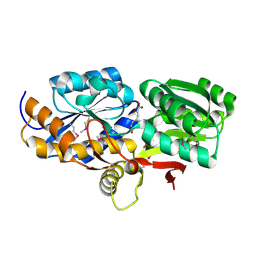 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with caffeic acid/3-(4-HYDROXY-PHENYL)PYRUVIC ACID | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, CAFFEIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4F06
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 RPB_2270 in complex with P-HYDROXYBENZOIC ACID | | Descriptor: | Extracellular ligand-binding receptor, GLYCEROL, P-HYDROXYBENZOIC ACID, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-15 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
3UKJ
 
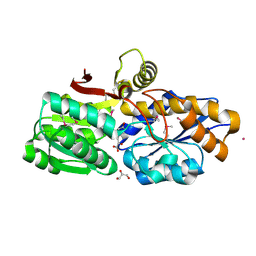 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3UK0
 
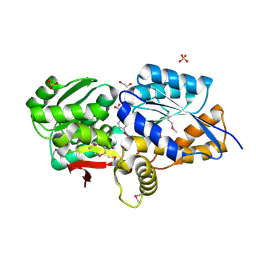 | | RPD_1889 protein, an extracellular ligand-binding receptor from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Osipiuk, J, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
2W27
 
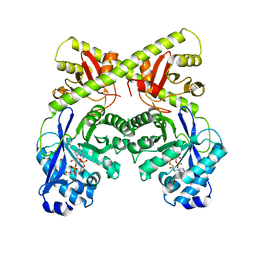 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | Authors: | Padavattan, S, AndERSON, W.F, Schirmer, T. | | Deposit date: | 2008-10-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
3SG0
 
 | | The crystal structure of an extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | BENZOYL-FORMIC ACID, Extracellular ligand-binding receptor | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3RPW
 
 | | The crystal structure of an ABC transporter from Rhodopseudomonas palustris CGA009 | | Descriptor: | ABC transporter, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4DQD
 
 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4EY3
 
 | | Crystal structure of solute binding protein of ABC transporter in complex with p-hydroxybenzoic acid | | Descriptor: | P-HYDROXYBENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4EYG
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with vanillic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXY-3-METHOXYBENZOATE, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4EYK
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with 3,4-dihydroxy benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4EYO
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4F8J
 
 | | The structure of an aromatic compound transport protein from Rhodopseudomonas palustris in complex with p-coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-17 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4FB4
 
 | | The Structure of an ABC-Transporter Family Protein from Rhodopseudomonas palustris in Complex with Caffeic Acid | | Descriptor: | CAFFEIC ACID, GLYCEROL, Putative branched-chain amino acid transport system substrate-binding protein | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-22 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4I1D
 
 | | The crystal structure of an ABC transporter substrate-binding protein from Bradyrhizobium japonicum USDA 110 | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, D-MALATE, ... | | Authors: | Fan, Y, Tan, K, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
1NSL
 
 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
2O6I
 
 | | Structure of an Enterococcus Faecalis HD Domain Phosphohydrolase | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, HD domain protein, ... | | Authors: | Vorontsov, I.I, Minasov, G, Shuvalova, L, Brunzelle, J.S, Moy, S, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex.
J.Biol.Chem., 286, 2011
|
|
