2G7Z
 
 | | Conserved DegV-like Protein of Unknown Function from Streptococcus pyogenes M1 GAS Binds Long-chain Fatty Acids | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, ... | | Authors: | Nocek, B, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conserved hypothetical protein from Streptococcus pyogenes M1 GAS discloses long-fatty acid (heptadecanoic acid) binding function
To be Published
|
|
4R23
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | Descriptor: | (3R,4R,5R)-3-(2,6-dichlorophenyl)-N-{(1R)-1-[(2R,4S)-4-(dihydroxymethyl)-5,5-dimethyl-1,3-thiazolidin-2-yl]-2-oxoethyl} -5-methyl-1,2-oxazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
2G2C
 
 | |
4RGP
 
 | | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans | | Descriptor: | CALCIUM ION, Csm6_III-A, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans
To be Published
|
|
4RJ0
 
 | |
4RIZ
 
 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-07 | | Release date: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
2GNP
 
 | | Structural Genomics, the crystal structure of a transcriptional regulator from Streptococcus pneumoniae TIGR4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, transcriptional regulator | | Authors: | Tan, K, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a transcriptional regulator from Streptococcus pneumoniae
TIGR4
To be Published
|
|
2GS5
 
 | |
4RM1
 
 | |
2GS8
 
 | | Structure of mevalonate pyrophosphate decarboxylase from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, mevalonate pyrophosphate decarboxylase | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of mevalonate pyrophosphate decarboxylase from Streptococcus pyogenes
To be Published
|
|
4S17
 
 | | The crystal structure of glutamine synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, Glutamine synthetase, MAGNESIUM ION | | Authors: | Cuff, M, Tan, K, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-08 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of glutamine synthetase from Bifidobacterium adolescentis ATCC 15703
To be Published
|
|
4RW0
 
 | | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008 | | Descriptor: | GLYCEROL, Lipolytic protein G-D-S-L family, SODIUM ION | | Authors: | Nocek, B, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-30 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008
To be Published
|
|
4RWE
 
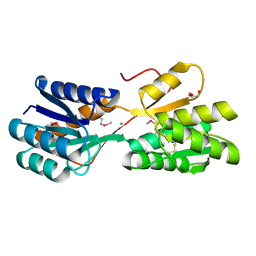 | | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, GLYCEROL, Sugar-binding transport protein | | Authors: | Tan, K, Zhou, M, Clancy, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-03 | | Release date: | 2014-12-31 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92
To be Published
|
|
4RLG
 
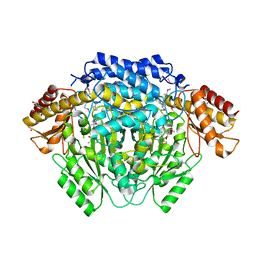 | |
2HAY
 
 | | The Crystal Structure of the Putative NAD(P)H-Flavin Oxidoreductase from Streptococcus pyogenes M1 GAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative NAD(P)H-flavin oxidoreductase, SULFATE ION | | Authors: | Kim, Y, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Crystal Structure of the Putative NAD(P)H-Flavin Oxidoreductase from Streptococcus pyogenes M1 GAS
To be Published, 2006
|
|
4U4E
 
 | |
2HNG
 
 | | The Crystal Structure of Protein of Unknown Function SP1558 from Streptococcus pneumoniae | | Descriptor: | Hypothetical protein | | Authors: | Kim, Y, Zhang, D, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-12 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Crystal Structure of Hypothetical Protein SP_1558 from Streptococcus pneumoniae
To be Published, 2006
|
|
2I1S
 
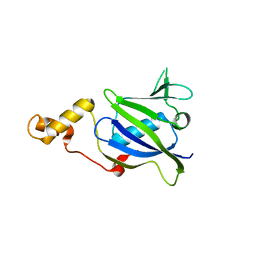 | |
2I5E
 
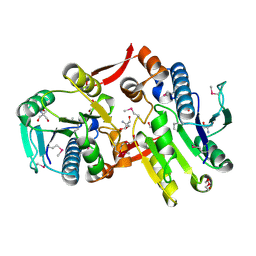 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
4O2H
 
 | | Crystal structure of BCAM1869 protein (RsaM homolog) from Burkholderia cenocepacia | | Descriptor: | protein BCAM1869 | | Authors: | Michalska, K, Chhor, G, Clancy, S, Winans, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-22 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RsaM: a transcriptional regulator of Burkholderia spp. with novel fold.
Febs J., 281, 2014
|
|
2OTD
 
 | | The crystal structure of the glycerophosphodiester phosphodiesterase from Shigella flexneri 2a | | Descriptor: | Glycerophosphodiester phosphodiesterase, PHOSPHATE ION | | Authors: | Zhang, R, Wu, R, Clancy, S, Jiang, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the glycerophosphodiester phosphodiesterase from Shigella flexneri 2a
To be Published
|
|
2R2A
 
 | | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Patterson, S, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-24 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis.
To be Published
|
|
3C8G
 
 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
3RQ1
 
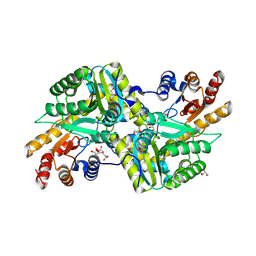 | | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula | | Descriptor: | 2-OXOGLUTARIC ACID, Aminotransferase class I and II, CHLORIDE ION, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Aminotransferase Class I and II from Veillonella parvula
To be Published
|
|
