3UTB
 
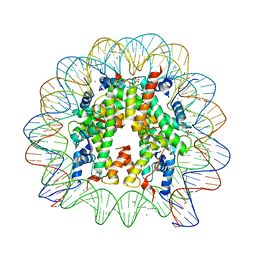 | | Crystal Structure of Nucleosome Core Particle Assembled with the 146b Alpha-Satellite Sequence (NCP146b) | | Descriptor: | 146-mer DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3UT9
 
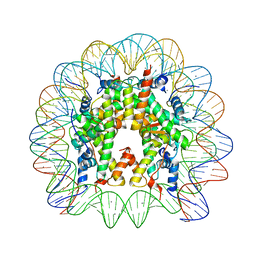 | | Crystal Structure of Nucleosome Core Particle Assembled with a Palindromic Widom '601' Derivative (NCP-601L) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3UTA
 
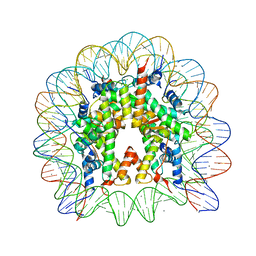 | | Crystal Structure of Nucleosome Core Particle Assembled with an Alpha-Satellite Sequence Containing Two TTAAA elements (NCP-TA2) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
6GEJ
 
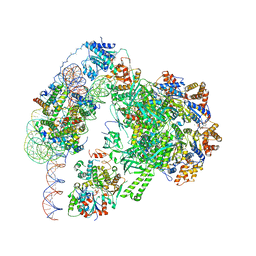 | | Chromatin remodeller-nucleosome complex at 3.6 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
8DUT
 
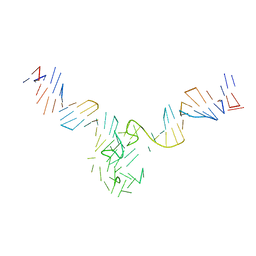 | | Duplex-G-quadruplex-duplex (DGD) class_1 | | Descriptor: | Duplex-G-quadruplex-duplex (46-MER) | | Authors: | Monsen, R.C, Chua, E.Y.D, Hopkins, J.B, Chaires, J.B, Trent, J.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 angstrom resolution with cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
6GEN
 
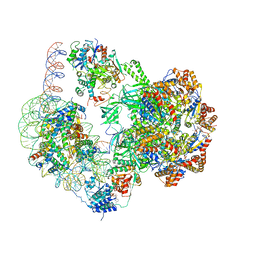 | | Chromatin remodeller-nucleosome complex at 4.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
4WU8
 
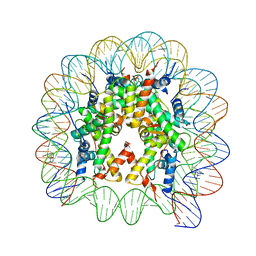 | | Structure of trPtNAP-NCP145 | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Stereochemical control of nucleosome targeting by platinum-intercalator antitumor agents.
Nucleic Acids Res., 43, 2015
|
|
4WU9
 
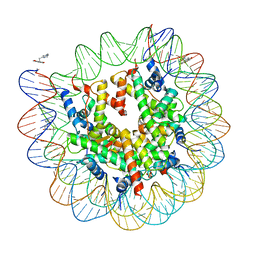 | | Structure of cisPtNAP-NCP145 | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Davey, G.E, Chin, C.F, Droge, P, Ang, W.H, Davey, C.A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stereochemical control of nucleosome targeting by platinum-intercalator antitumor agents.
Nucleic Acids Res., 43, 2015
|
|
9FBW
 
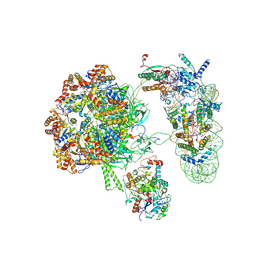 | | SWR1 lacking Swc5 subunit in complex with hexasome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2024-05-14 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
3LZ0
 
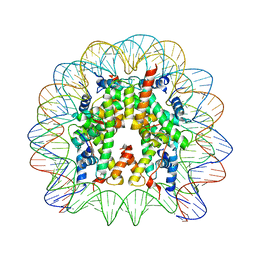 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 1) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LZ1
 
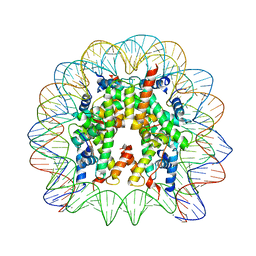 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 2) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
8QZ0
 
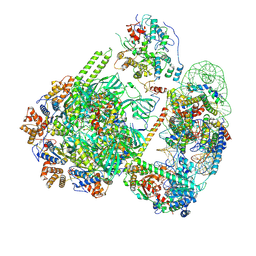 | | SWR1-hexasome-dimer complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
8QYV
 
 | | SWR1-hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
7UO7
 
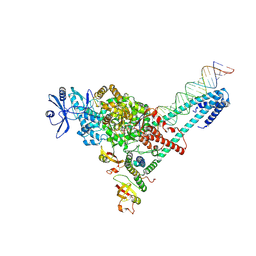 | | SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO4
 
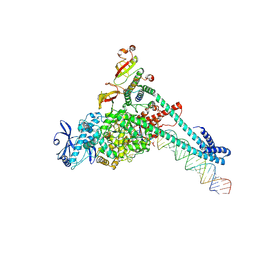 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOE
 
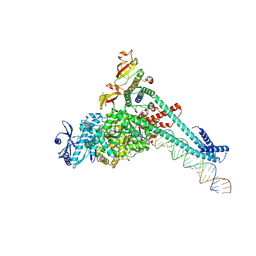 | | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
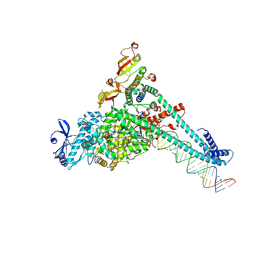 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
