7P63
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P69
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7L
 
 | | Complex I from E. coli, DDM/LMNG-purified, with NADH and FMN, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P62
 
 | | Complex I from E. coli, DDM-purified, Apo, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P61
 
 | | Complex I from E. coli, DDM-purified, with NADH, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P64
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7C
 
 | | Complex I from E. coli, DDM/LMNG-purified, Apo, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7E
 
 | | Complex I from E. coli, DDM/LMNG-purified, Apo, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7J
 
 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7M
 
 | | Complex I from E. coli, DDM/LMNG-purified, inhibited by Piericidin A, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7K
 
 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7T
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z83
 
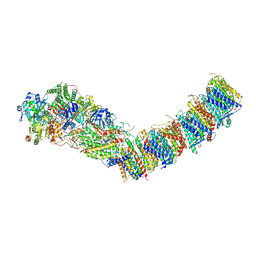 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z84
 
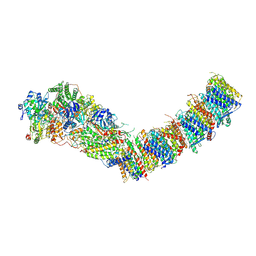 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7S
 
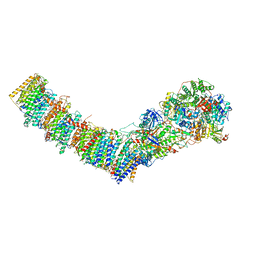 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZC5
 
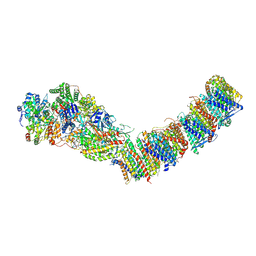 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z80
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7V
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7R
 
 | | Complex I from E. coli, LMNG-purified, Apo, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZCI
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-28 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
1SWG
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-07-12 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
1SWF
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 | | Descriptor: | CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-04-23 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
4GD9
 
 | | Circular Permuted Streptavidin N49/G48 | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Le Trong, I, Chu, V, Xing, Y, Lybrand, T.P, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural consequences of cutting a binding loop: two circularly permuted variants of streptavidin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GDA
 
 | | Circular Permuted Streptavidin A50/N49 | | Descriptor: | BIOTIN, ETHANOL, GLYCEROL, ... | | Authors: | Le Trong, I, Chu, V, Xing, Y, Lybrand, T.P, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural consequences of cutting a binding loop: two circularly permuted variants of streptavidin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1N43
 
 | | Streptavidin Mutant N23A with biotin at 1.89A | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
