7U6L
 
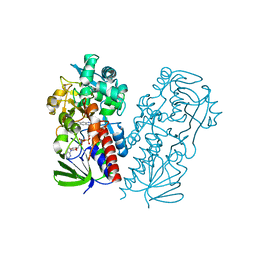 | | Pseudooxynicotine amine oxidase | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choudhary, V, Stull, F, Wu, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
9CFJ
 
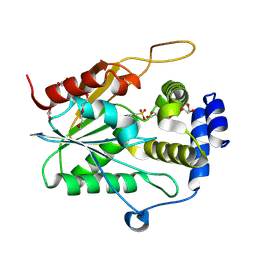 | | Fluvirucin Thioesterase Domain (FluC TE) | | Descriptor: | FluC, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Choudhary, V, Smith, J.L. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
5TYT
 
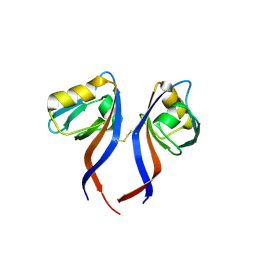 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
9CGN
 
 | | Pikromycin Thioesterase with heptaketide adduct | | Descriptor: | (2S,4R,5S,6S,8R,12R,13R)-5,13-dihydroxy-2,4,6,8,12-pentamethyl-3,9-dioxopentadecanal, Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | Smith, J.L, Choudhary, V. | | Deposit date: | 2024-06-30 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
9CEL
 
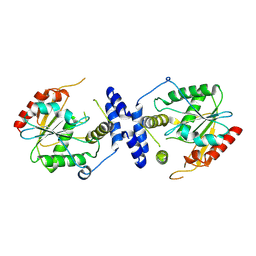 | | Juvenimicin Thioesterase | | Descriptor: | Type I PKS module 7 | | Authors: | Akey, D.L, Smith, J.S, Choudhary, V. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
9CGO
 
 | | Tylosin thioesterase domain (TylG5 TE) | | Descriptor: | Tylactone synthase module 7 | | Authors: | Smith, J.L, Choudhary, V. | | Deposit date: | 2024-06-30 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
7TLX
 
 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
9CBD
 
 | | Pikromycin Thioesterase Domain | | Descriptor: | Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
9CGL
 
 | | Pikromycin Thioesterase Doubly Protected DAP | | Descriptor: | 2-{[(1R)-1-(6-nitro-2H-1,3-benzodioxol-5-yl)ethyl]sulfanyl}ethyl formate, Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2024-06-29 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
