4AII
 
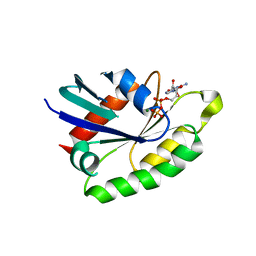 | | Crystal structure of the rat REM2 GTPase - G domain bound to GDP | | Descriptor: | GTP-BINDING PROTEIN REM 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Reymond, P, Coquard, A, Chenon, M, Zeghouf, M, El Marjou, A, Thompson, A, Menetrey, J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Gdp-Bound G Domain of the Rgk Protein Rem2.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5OJ8
 
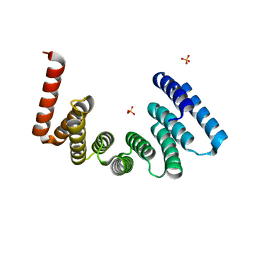 | | Crystal structure of the KLC1-TPR domain ([A1-B5] fragment) | | Descriptor: | Kinesin light chain 1, PHOSPHATE ION | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
5OJF
 
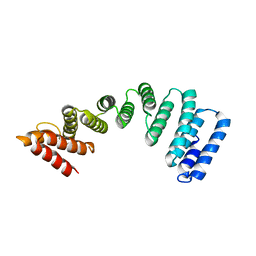 | | Crystal Structure of KLC2-TPR domain (fragment [A1-B6] | | Descriptor: | Kinesin light chain 2 | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Andreani, J, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
1I5K
 
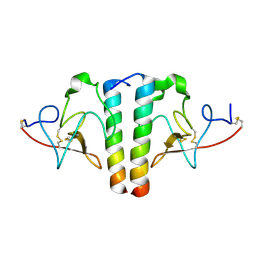 | | STRUCTURE AND BINDING DETERMINANTS OF THE RECOMBINANT KRINGLE-2 DOMAIN OF HUMAN PLASMINOGEN TO AN INTERNAL PEPTIDE FROM A GROUP A STREPTOCOCCAL SURFACE PROTEIN | | Descriptor: | M PROTEIN, PLASMINOGEN | | Authors: | Rios-Steiner, J.L, Schenone, M, Mochalkin, I, Tulinsky, A, Castellino, F.J. | | Deposit date: | 2001-02-27 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein.
J.Mol.Biol., 308, 2001
|
|
4A5U
 
 | |
4PXJ
 
 | |
7A3Z
 
 | | OSM-3 kinesin motor domain complexed with Mg.ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Osmotic avoidance abnormal protein 3 | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
7A40
 
 | | Nucleotide-free OSM-3 kinesin motor domain | | Descriptor: | GLYCEROL, Osmotic avoidance abnormal protein 3, SULFATE ION | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
7A5E
 
 | | OSM-3 kinesin motor domain complexed with Mg.AMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, Osmotic avoidance abnormal protein 3, ... | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
6EW6
 
 | | Crystal structure of the BCL6 BTB domain in complex with anilinopyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}2-(2-chlorophenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6.
ACS Chem. Biol., 13, 2018
|
|
6EW8
 
 | |
6EW7
 
 | |
