4X36
 
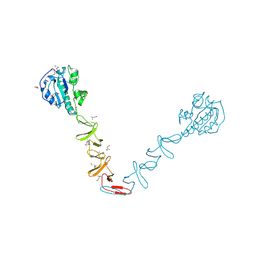 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XSP
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP | | Descriptor: | Alr3699 protein, GLYCEROL, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSR
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP-glucose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alr3699 protein, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSO
 
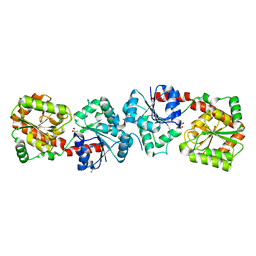 | | Crystal structure of apo-form Alr3699/HepE from Anabaena sp. strain PCC 7120 | | Descriptor: | Alr3699 protein, GLYCEROL | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSU
 
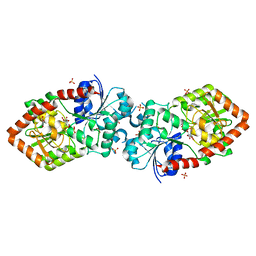 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP and glucose | | Descriptor: | Alr3699 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4NCD
 
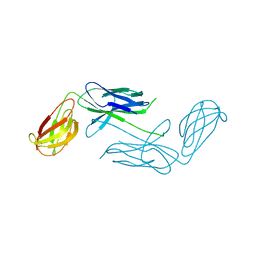 | | Crystal Structure of Class 5 Fimbriae Chaperone CfaA | | Descriptor: | Gram-negative pili assembly chaperone, N-terminal domain protein | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae.
Plos Pathog., 10, 2014
|
|
4OBX
 
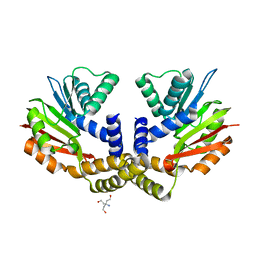 | | Crystal structure of yeast Coq5 in the apo form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OBW
 
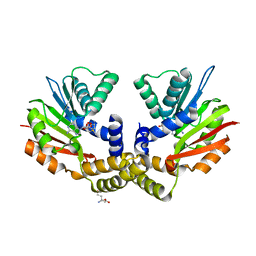 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3D8X
 
 | | Crystal Structure of Saccharomyces cerevisiae NDPPH Dependent Thioredoxin Reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Zhang, Z.Y, Bao, R, Yu, J, Chen, Y.X, Zhou, C.-Z. | | Deposit date: | 2008-05-26 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin
Biochim.Biophys.Acta, 1794, 2009
|
|
4JHP
 
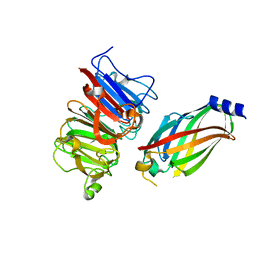 | | The crystal structure of the RPGR RCC1-like domain in complex with PDE6D | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, X-linked retinitis pigmentosa GTPase regulator | | Authors: | Waetzlich, D, Vetter, I, Wittinghofer, A, Ismail, S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The interplay between RPGR, PDE-delta and Arl2/3 regulate the ciliary targeting of farnesylated cargo.
Embo Rep., 14, 2013
|
|
4JHN
 
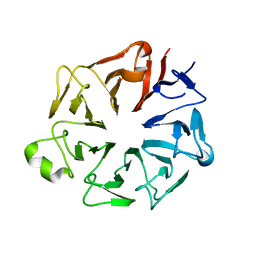 | | The crystal structure of the RPGR RCC1-like domain | | Descriptor: | X-linked retinitis pigmentosa GTPase regulator | | Authors: | Waetzlich, D, Vetter, I, Wittinghofer, A, Ismail, S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The interplay between RPGR, PDE-delta and Arl2/3 regulate the ciliary targeting of farnesylated cargo.
Embo Rep., 14, 2013
|
|
7VZO
 
 | |
3QWB
 
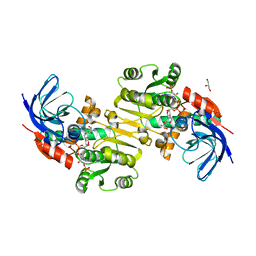 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 complexed with NADPH | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QWA
 
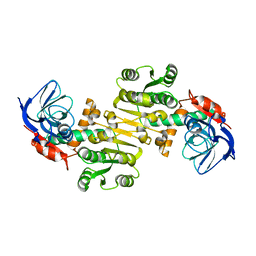 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 | | Descriptor: | Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
4IPN
 
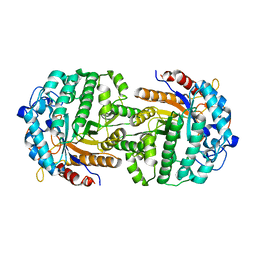 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4IPL
 
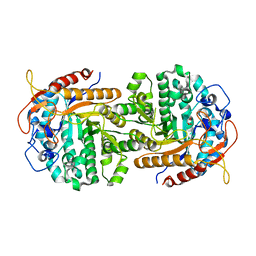 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
5Y4U
 
 | | Crystal structure of Grx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | Monothiol glutaredoxin-3 | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5Y4T
 
 | | Crystal structure of Trx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutaredoxin | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5Y4B
 
 | | Solution structure of yeast Fra2 | | Descriptor: | BolA-like protein 2 | | Authors: | Tang, Y.J, Chi, C.B, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J. Mol. Biol., 430, 2018
|
|
7YUK
 
 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
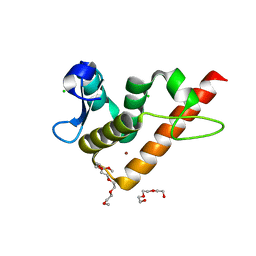 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
