6UGJ
 
 | |
6UGG
 
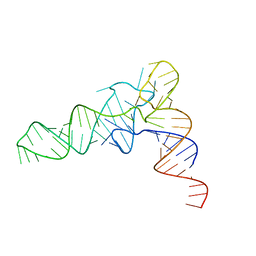 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
6UGI
 
 | |
6VMY
 
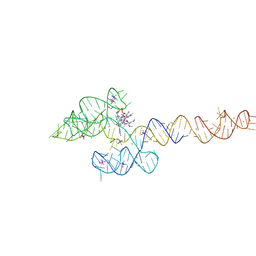 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
6CWX
 
 | | Crystal structure of human ribonuclease P/MRP proteins Rpp20/Rpp25 | | Descriptor: | FORMIC ACID, Ribonuclease P protein subunit p20, Ribonuclease P protein subunit p25, ... | | Authors: | Chan, C.W, Kiesel, B.R, Mondragon, A. | | Deposit date: | 2018-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rpp20/Rpp25 Reveals Quaternary Level Adaptation of the Alba Scaffold as Structural Basis for Single-stranded RNA Binding.
J. Mol. Biol., 430, 2018
|
|
7LJP
 
 | | Structure of Thermotoga maritima SmpB | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2021-01-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Thermotoga maritima SmpB reveals its C-terminal tail domain in a helical conformation mimicking that of a ribosome-bound state
To Be Published
|
|
4ZQA
 
 | |
1XGE
 
 | | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between subunits | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Chan, C.W, Guss, J.M, Christopherson, R.I, Maher, M.J. | | Deposit date: | 2004-09-17 | | Release date: | 2005-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between Subunits
J.Mol.Biol., 348, 2005
|
|
5D90
 
 | |
5D8C
 
 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to promoter DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*AP*AP*CP*TP*CP*TP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*TP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3'), MerR family regulator protein | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
5E01
 
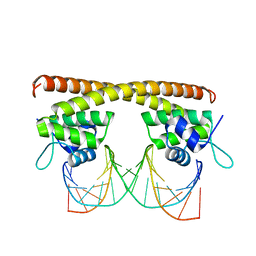 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to palyndromic promoter DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3', Uncharacterized HTH-type transcriptional regulator HI_0186 | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-09-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
4BZ1
 
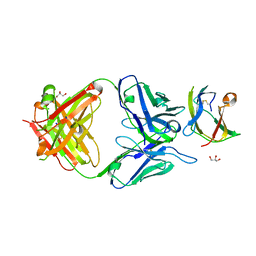 | | Structure of dengue virus EDIII in complex with Fab 3e31 | | Descriptor: | CHLORIDE ION, ENVELOPE PROTEIN, FAB 3E31 HEAVY CHAIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 3E31
To be Published
|
|
4BZ2
 
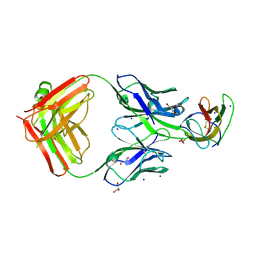 | | Structure of dengue virus EDIII in complex with Fab 2D73 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ENVELOPE PROTEIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 2D73
To be Published
|
|
4ZM1
 
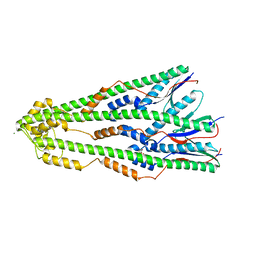 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - wild type | | Descriptor: | CITRIC ACID, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
4ZM5
 
 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - A107P mutant | | Descriptor: | CHLORIDE ION, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
2EG8
 
 | | The crystal structure of E. coli dihydroorotase complexed with 5-fluoroorotic acid | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
2EG6
 
 | |
2EG7
 
 | | The crystal structure of E. coli dihydroorotase complexed with HDDP | | Descriptor: | 2-OXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4,6-DICARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
3P0U
 
 | | Crystal Structure of the ligand binding domain of human testicular receptor 4 | | Descriptor: | Nuclear receptor subfamily 2 group C member 2 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Xu, Y, Chan, C.-W, Kruse, S.W, Reynolds, R, Engel, J.D, Xu, H.E. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Orphan Nuclear Receptor TR4 Is a Vitamin A-activated Nuclear Receptor.
J.Biol.Chem., 286, 2011
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
8IO9
 
 | |
8IOA
 
 | |
8IO7
 
 | |
8IO6
 
 | |
8IO8
 
 | |
