6H1C
 
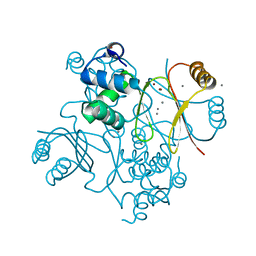 | | Structure of Ferric uptake regulator from Pseudomonas aeruginosa with manganese. | | Descriptor: | Ferric uptake regulation protein, MANGANESE (II) ION, ZINC ION | | Authors: | Nader, S, Perard, J, Carpentier, P, Michaud-Soret, I, Crouzy, S. | | Deposit date: | 2018-07-11 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ferric uptake regulator with Mn and Zn form Pseudomonas aeruginosa
To Be Published
|
|
6HK5
 
 | | X-ray structure of a truncated mutant of the metallochaperone CooJ with a high-affinity nickel-binding site | | Descriptor: | 3,3',3''-phosphoryltripropanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Alfano, M, Perard, J, Basset, C, Carpentier, P, Zambelli, B, Timm, J, Crouzy, S, Ciurli, S, Cavazza, C. | | Deposit date: | 2018-09-05 | | Release date: | 2019-03-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | The carbon monoxide dehydrogenase accessory protein CooJ is a histidine-rich multidomain dimer containing an unexpected Ni(II)-binding site.
J.Biol.Chem., 294, 2019
|
|
5O17
 
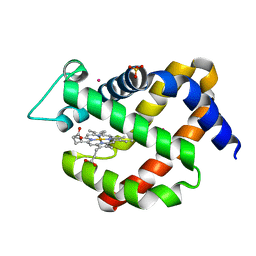 | | Crystal structure of murine neuroglobin under 100 bar krypton | | Descriptor: | KRYPTON, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Colloc'h, N, Prange, T, Vallone, B, Carpentier, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping Hydrophobic Tunnels and Cavities in Neuroglobin with Noble Gas under Pressure.
Biophys. J., 113, 2017
|
|
7PWN
 
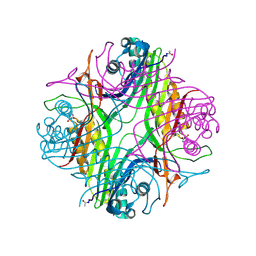 | | URATE OXYDASE AZA-XANTHINE COMPLEX AT 1000 BARS (100 MPa) OF ARGON | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
7Q09
 
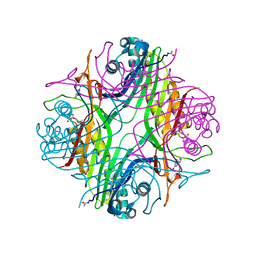 | | URATE OXIDASE AZA-XANTHINE COMPLEX UNDER 1500 BAR OF ARGON | | Descriptor: | 8-AZAXANTHINE, ACETYL GROUP, ARGON, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
4TTT
 
 | |
6I9X
 
 | | urate oxidase under 35 bar of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
3TMT
 
 | | IrisFP, distorted chromophore | | Descriptor: | Green to red photoconvertible GPF-like protein EosFP, SULFATE ION | | Authors: | Adam, V, Carpentier, P, Roy, A, Field, M, Bourgeois, D. | | Deposit date: | 2011-08-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nature of transient dark States in a photoactivatable fluorescent protein.
J.Am.Chem.Soc., 133, 2011
|
|
8ORO
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER HYPEROXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-04-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8OXN
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) S101A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXYC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8OXT
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) H251A VARIANT COMPLEXED WITH N-ACETYLANTHRANILATE AS RESULT OF IN CRYSTALLO TURNOVER OF ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-(ACETYLAMINO)BENZOIC ACID, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8A97
 
 | | ROOM TEMPERATURE CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) UNDER XENON PRESSURE (30 bar) | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, D(-)-TARTARIC ACID, XENON | | Authors: | Bui, S, Prange, T, Steiner, R.A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
3DVW
 
 | |
3DVX
 
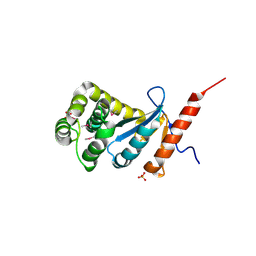 | | Crystal structure of reduced DsbA3 from Neisseria meningitidis | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Lafaye, C, Serre, L. | | Deposit date: | 2008-07-21 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis.
J.Mol.Biol., 392, 2009
|
|
5MDL
 
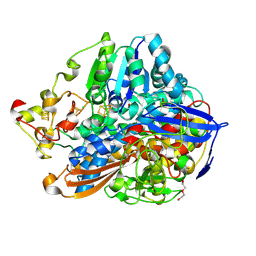 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its O2-derivatized form by a "soak-and-freeze" derivatization method | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MDK
 
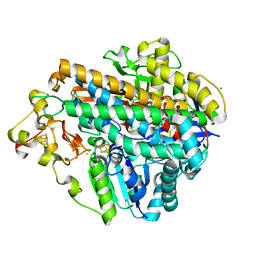 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form (oxidized state - state 3) | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MDJ
 
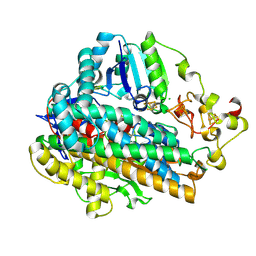 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in a its as-isolated high-pressurized form | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2B3Y
 
 | | Structure of a monoclinic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
2B3X
 
 | | Structure of an orthorhombic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, Iron-responsive element binding protein 1, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
6SWV
 
 | | Trypsin fast data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | von Stetten, D, Mueller-Dieckmann, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | ID30A-3 (MASSIF-3) - a beamline for macromolecular crystallography at the ESRF with a small intense beam.
J.Synchrotron Radiat., 27, 2020
|
|
6I3T
 
 | | Crystal structure of murine neuroglobin bound to CO at 40 K. | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C. | | Deposit date: | 2018-11-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
6XYT
 
 | | Crystal structure of the O-state of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC2
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ALANINE, EICOSANE, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
