9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
5IO9
 
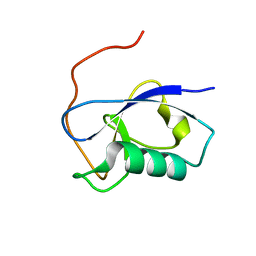 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IP4
 
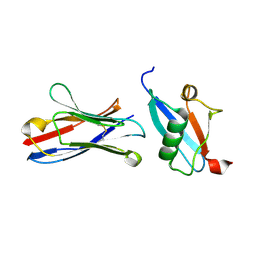 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IN7
 
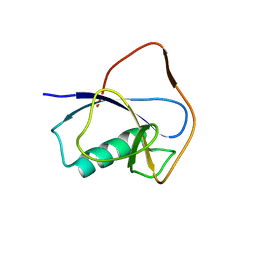 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IOI
 
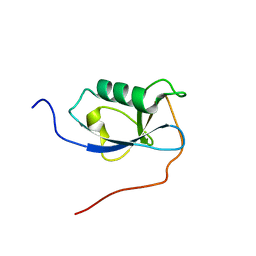 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5QU1
 
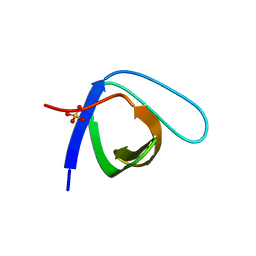 | | Crystal Structure of the monomeric human Nck SH3.1 domain, triclinic, 1.08A | | Descriptor: | Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU5
 
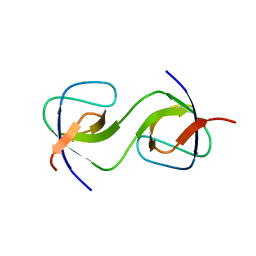 | | Domain Swap in the first SH3 domain of human Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
3AQ1
 
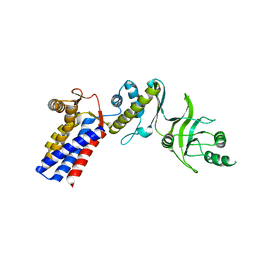 | | Open state monomer of a group II chaperonin from methanococcoides burtonii | | Descriptor: | Thermosome subunit | | Authors: | Harrop, S.J, Pilak, O, Siddiqui, K.S, De Francisci, D, Burg, D, Williams, T.J, Cavicchioli, R, Curmi, P.M. | | Deposit date: | 2010-10-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Chaperonins from an Antarctic archaeon are predominantly monomeric: crystal structure of an open state monomer.
ENVIRON.MICROBIOL., 13, 2011
|
|
3KWX
 
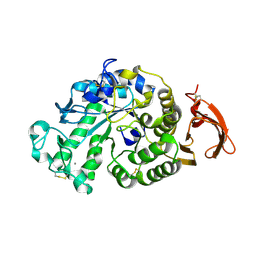 | | Chemically modified Taka alpha-amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase A type-1/2, CALCIUM ION | | Authors: | Siddiqui, K.S, Harrop, S.J, Poljak, A, De Francisci, D, Guerriero, G, Pilak, O, Burg, D, Raftery, M.J, Parkin, D.M, Trewhella, J, Cavicchioli, R. | | Deposit date: | 2009-12-01 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Modified alpha-amylase with a molten-globule state has enhanced thermal stability
To be Published
|
|
1QQ9
 
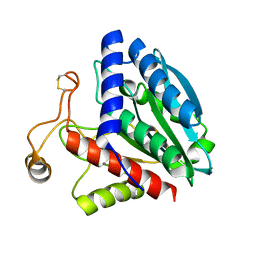 | | STREPTOMYCES GRISEUS AMINOPEPTIDASE COMPLEXED WITH METHIONINE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, METHIONINE, ... | | Authors: | Gilboa, R, Greenblatt, H.M, Perach, M, Spungin-Bialik, A, Lessel, U, Schomburg, D, Blumberg, S, Shoham, G. | | Deposit date: | 1999-06-12 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QW9
 
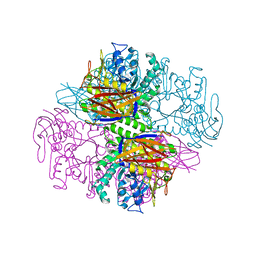 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | Descriptor: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QW8
 
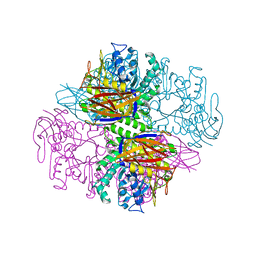 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
3ZKX
 
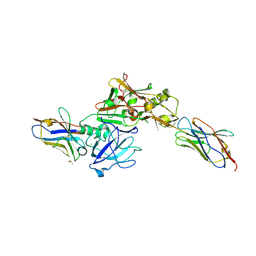 | | TERNARY BACE2 XAPERONE COMPLEX | | Descriptor: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1QI9
 
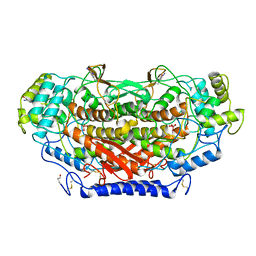 | | X-RAY SIRAS STRUCTURE DETERMINATION OF A VANADIUM-DEPENDENT HALOPEROXIDASE FROM ASCOPHYLLUM NODOSUM AT 2.0 A RESOLUTION | | Descriptor: | VANADATE ION, Vanadium-dependent bromoperoxidase | | Authors: | Weyand, M, Hecht, H.-J, Kiess, M, Liaud, M.F, Vilter, H, Schomburg, D. | | Deposit date: | 1999-06-10 | | Release date: | 2000-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution.
J.Mol.Biol., 293, 1999
|
|
3LIP
 
 | |
2JAE
 
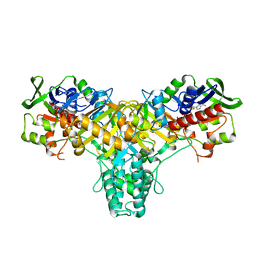 | | The structure of L-amino acid oxidase from Rhodococcus opacus in the unbound state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation.
J.Mol.Biol., 367, 2007
|
|
2JB3
 
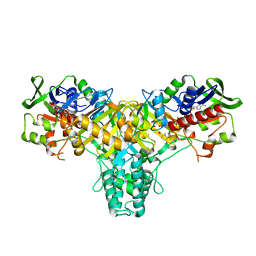 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB2
 
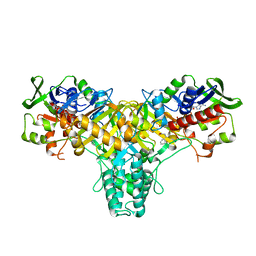 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with L-phenylalanine. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE, PHENYLALANINE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB1
 
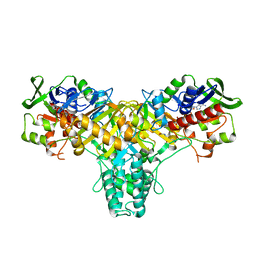 | | The L-amino acid oxidase from Rhodococcus opacus in complex with L- alanine | | Descriptor: | ALANINE, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
3H52
 
 | | Crystal structure of the antagonist form of human glucocorticoid receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Benz, J, D'Arcy, B, Stihle, M, Burger, D, Thoma, R, Ruf, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations
J.Mol.Biol., 395, 2010
|
|
2EXH
 
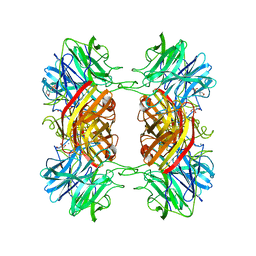 | | Structure of the family43 beta-Xylosidase from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Yuval, S, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
3MQE
 
 | | Structure of SC-75416 bound at the COX-2 active site | | Descriptor: | (2S)-7-tert-butyl-6-chloro-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Springer, J, Rogier, J, Kiefer, J.R. | | Deposit date: | 2010-04-28 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3JYP
 
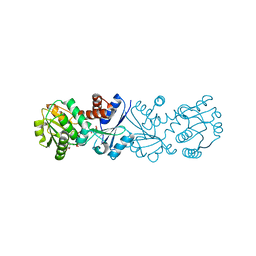 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYO
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Niefind, K, Schomburg, D. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3NTG
 
 | | Crystal structure of COX-2 with selective compound 23d-(R) | | Descriptor: | (2R)-6,8-dichloro-7-(2-methylpropoxy)-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Carter, J.C, Talley, J.J, Kiefer, J.R. | | Deposit date: | 2010-07-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
