9B22
 
 | |
9B21
 
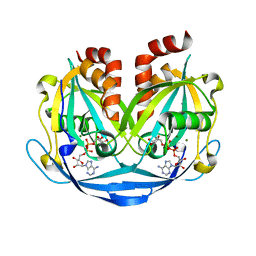 | | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form) | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-03-14 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form)
To be published
|
|
5KWX
 
 | |
5KWO
 
 | |
5KX2
 
 | |
5KWP
 
 | |
5KX0
 
 | |
5KX1
 
 | |
5KWZ
 
 | |
5KVN
 
 | |
6CKP
 
 | |
6NUP
 
 | |
6Q09
 
 | |
6PTG
 
 | |
6TYJ
 
 | |
6VWE
 
 | |
7S5O
 
 | |
4LGV
 
 | |
4O2D
 
 | |
5EJ2
 
 | |
3D63
 
 | |
3DAH
 
 | |
3EIZ
 
 | |
3EK2
 
 | |
3EJ2
 
 | |
