2KJD
 
 | |
2JXO
 
 | | Structure of the second PDZ domain of NHERF-1 | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Cheng, H, Li, J, Dai, Z, Bu, Z, Roder, H. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory Interactions between the PDZ2 and C-terminal Domains in the Scaffolding Protein NHERF1
Structure, 17, 2009
|
|
2KRG
 
 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
2J8J
 
 | | Solution Structure of the A4 Domain of Blood Coagulation Factor XI | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2J8L
 
 | | FXI Apple 4 domain loop-out conformation | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2M0U
 
 | | Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0V
 
 | | Complex structure of C-terminal CFTR peptide and extended PDZ2 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0T
 
 | |
4PLI
 
 | |
4PL6
 
 | |
4PLL
 
 | |
8YQV
 
 | | African swine fever virus RNA Polymerase core | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Transcription regulation of African swine fever virus: dual role of M1249L.
Nat Commun, 15, 2024
|
|
8YQU
 
 | |
8YQY
 
 | | ASFV RNA polymerase-M1249L complex complete | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Transcription regulation of African swine fever virus: dual role of M1249L.
Nat Commun, 15, 2024
|
|
8YQT
 
 | |
8YQZ
 
 | |
8YQX
 
 | | ASFV RNA polymerase-M1249L complex4 | | Descriptor: | D339L, DNA-directed RNA polymerase subunit, M1249L | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Transcription regulation of African swine fever virus: dual role of M1249L.
Nat Commun, 15, 2024
|
|
8YQW
 
 | | ASFV RNA polymerase-M1249L complex3 | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Transcription regulation of African swine fever virus: dual role of M1249L.
Nat Commun, 15, 2024
|
|
7CBT
 
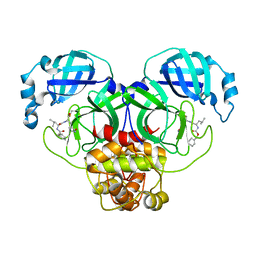 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Shi, Y, Peng, G. | | Deposit date: | 2020-06-13 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
6L2T
 
 | |
7WGX
 
 | | SARS-CoV-2 spike glycoprotein trimer in closed state after treatment with Cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
