6OB1
 
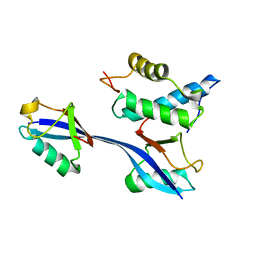 | | Structure of WHB in complex with Ubiquitin Variant | | Descriptor: | Anaphase-promoting complex subunit 2, Ubiquitin | | Authors: | Edmond, R.W, Grace, C.R. | | Deposit date: | 2019-03-19 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8AMZ
 
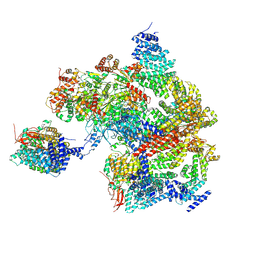 | | Spinach 19S proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1 homolog, 26S proteasome non-ATPase regulatory subunit 2 homolog, 26S proteasome regulatory subunit 7, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome
Plant Communications, 3, 2022
|
|
4PM6
 
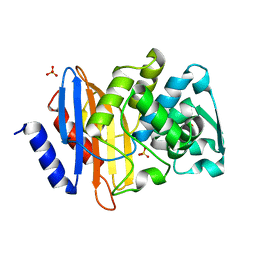 | | Crystal structure of CTX-M-14 S70G beta-lactamase at 1.56 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4RG6
 
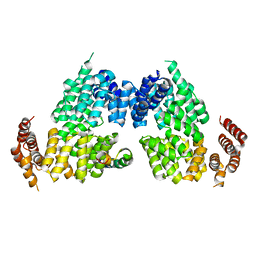 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4PM8
 
 | | Crystal structure of CTX-M-14 S70G:S237A beta-lactamase at 1.17 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.169 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4RG7
 
 | | Crystal structure of APC3 | | Descriptor: | Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4RG9
 
 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4PM7
 
 | | Crystal structure of CTX-M-14 S70G:S237A in complex with cefotaxime at 1.29 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM5
 
 | | Crystal structure of CTX-M-14 S70G beta-lactamase in complex with cefotaxime at 1.26 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
3OCP
 
 | | Crystal structure of cAMP bound cGMP-dependent protein kinase(92-227) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3OD0
 
 | | Crystal structure of cGMP bound cGMP-dependent protein kinase(92-227) | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
3OGJ
 
 | | Crystal structure of partial apo (92-227) of cGMP-dependent protein kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, PRKG1 protein | | Authors: | Kim, J.J, Huang, G, Kwon, T.K, Zwart, P, Headd, J, Kim, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Co-Crystal Structures of PKG Ibeta (92-227) with cGMP and cAMP Reveal the Molecular Details of Cyclic-Nucleotide Binding
Plos One, 6, 2011
|
|
