3PND
 
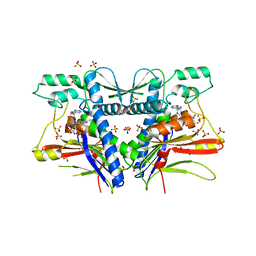 | | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD binding proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Boyd, J.M, Endrizzi, J.A, Hamilton, T.L, Christopherson, M.R, Mulder, D.W, Downs, D.M, Peters, J.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins.
J.Bacteriol., 193, 2011
|
|
6KVQ
 
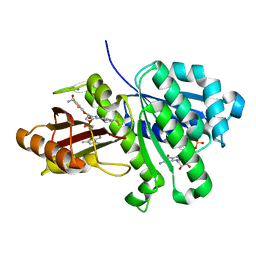 | | S. aureus FtsZ in complex with BOFP (compound 3) | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
6KVP
 
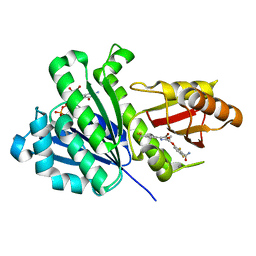 | | S. aureus FtsZ in complex with 3-(1-(5-bromo-4-(4-(trifluoromethyl)phenyl)oxazol-2-yl)ethoxy)-2,6-difluorobenzamide (compound 2) | | Descriptor: | 3-[(1R)-1-[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]ethoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
6PAJ
 
 | |
8D8S
 
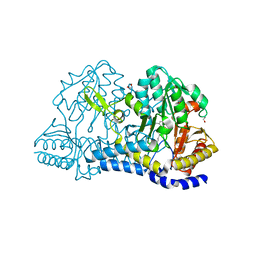 | | SufS from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase | | Authors: | Morrison, C.N, Boncella, A.E, Lunin, V. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Structural and Biochemical Characterization of Staphylococcus aureus Cysteine Desulfurase Complex SufSU.
Acs Omega, 7, 2022
|
|
2KY9
 
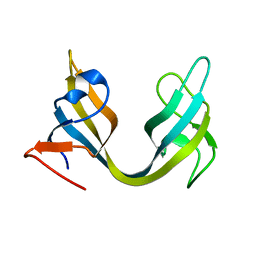 | | Solution NMR Structure of ydhK C-terminal Domain from B.subtilis, Northeast Structural Genomics Consortium Target Target SR518 | | Descriptor: | Uncharacterized protein ydhK | | Authors: | Eletsky, A, Sukumaran, D.K, Lee, H, Lee, D, Ciccosanti, C, Janjua, H, Liu, J, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-21 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The copBL operon protects Staphylococcus aureus from copper toxicity: CopL is an extracellular membrane-associated copper-binding protein.
J.Biol.Chem., 294, 2019
|
|
