3I55
 
 | | Co-crystal structure of Mycalamide A Bound to the Large Ribosomal Subunit | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | 著者 | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | 登録日 | 2009-07-03 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
3I56
 
 | | Co-crystal structure of Triacetyloleandomcyin Bound to the Large Ribosomal Subunit | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | 著者 | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | 登録日 | 2009-07-03 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
2QEX
 
 | |
4ZCK
 
 | | Crystal Structure of C-terminal Fragment of Escherichia coli BipA/TypA | | 分子名称: | GTP-binding protein TypA/BipA, MAGNESIUM ION | | 著者 | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | 登録日 | 2015-04-16 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZCM
 
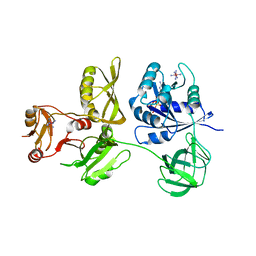 | | Crystal Structure of Escherichia coli GTPase BipA/TypA Complexed with ppGpp | | 分子名称: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | 著者 | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | 登録日 | 2015-04-16 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZCL
 
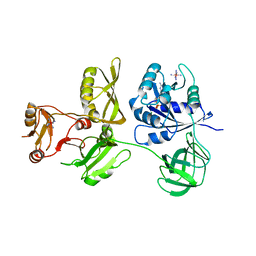 | | Crystal Structure of Escherichia coli GTPase BipA/TypA Complexed with GDP | | 分子名称: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | 登録日 | 2015-04-16 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZCI
 
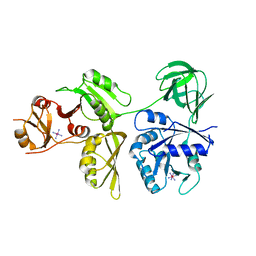 | | Crystal Structure of Escherichia coli GTPase BipA/TypA | | 分子名称: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA | | 著者 | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | 登録日 | 2015-04-16 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.627 Å) | | 主引用文献 | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
1YI2
 
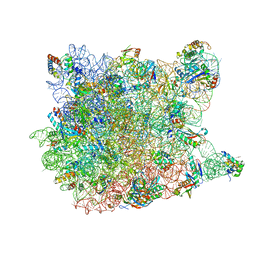 | | Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-11 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YHQ
 
 | | Crystal Structure Of Azithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-10 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIJ
 
 | | Crystal Structure Of Telithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-12 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJ9
 
 | | Crystal Structure Of The Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui Containing a three residue deletion in L22 | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-13 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJN
 
 | | Crystal Structure Of Clindamycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-14 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJW
 
 | | Crystal Structure Of Quinupristin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S RIBOSOMAL RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-15 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIT
 
 | | Crystal Structure Of Virginiamycin M and S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-13 | | 公開日 | 2005-04-26 | | 最終更新日 | 2012-12-12 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
3LZJ
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dCTP Opposite 7,8-Dihydro-8-oxoguanine | | 分子名称: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | 著者 | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | 登録日 | 2010-03-01 | | 公開日 | 2010-05-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
3LZI
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dATP Opposite 7,8-dihydro-8-oxoguanine | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | 著者 | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | 登録日 | 2010-03-01 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
3NCI
 
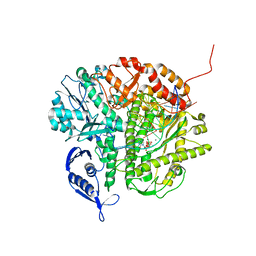 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite dG at 1.8 angstrom resolution | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | 著者 | Wang, M, Blaha, G, Steitz, T.A, Konigsberg, W.H, Wang, J. | | 登録日 | 2010-06-04 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex.
Biochemistry, 50, 2011
|
|
3NAE
 
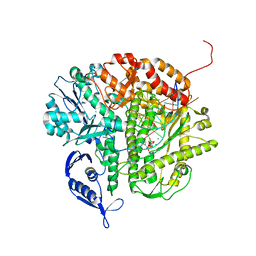 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite Guanidinohydantoin | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | 著者 | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | 登録日 | 2010-06-01 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Substitution of Ala for Tyr567 in RB69 DNA Polymerase Allows dAMP and dGMP To Be Inserted opposite Guanidinohydantoin .
Biochemistry, 49, 2010
|
|
3CB4
 
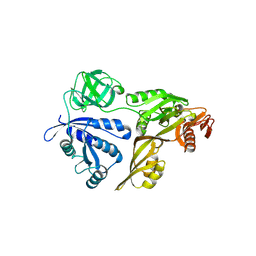 | | The Crystal Structure of LepA | | 分子名称: | GTP-binding protein lepA | | 著者 | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | 登録日 | 2008-02-21 | | 公開日 | 2008-03-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7JLT
 
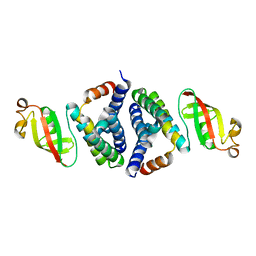 | |
1P9Y
 
 | |
1OMS
 
 | |
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X7K
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
