7UZL
 
 | |
6U1S
 
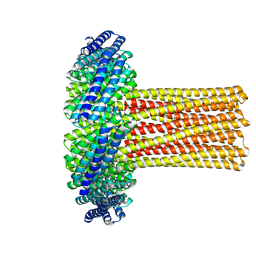 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | 分子名称: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | 著者 | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | 登録日 | 2019-08-16 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Computational Design of Transmembrane Channels
To Be Published
|
|
6WXP
 
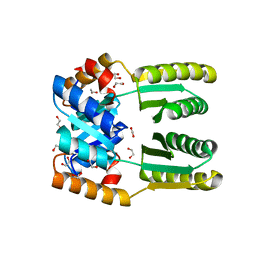 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 4-glutamate centre TFD-EE | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, TFD-EE | | 著者 | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | 登録日 | 2020-05-11 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WXO
 
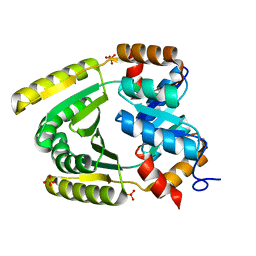 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 2-histidine 2-glutamate centre TFD-HE | | 分子名称: | GLYCEROL, SULFATE ION, TFD-HE | | 著者 | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | 登録日 | 2020-05-11 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | 分子名称: | C4_nat_HFuse-7900 | | 著者 | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | 登録日 | 2020-07-16 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XI6
 
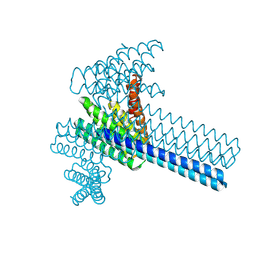 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | 分子名称: | helical fusion design | | 著者 | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | 登録日 | 2020-06-19 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XH5
 
 | |
8CUN
 
 | |
8CWA
 
 | |
8CTO
 
 | |
8CUX
 
 | |
8CUV
 
 | |
8CUT
 
 | |
8CUS
 
 | |
8CWZ
 
 | |
8CUU
 
 | |
8CWY
 
 | |
8CWS
 
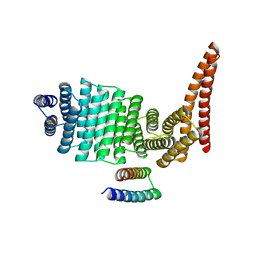 | |
8CUW
 
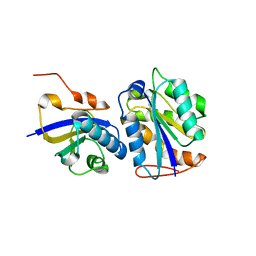 | |
7UBD
 
 | |
7UBC
 
 | |
7UBF
 
 | |
7UCP
 
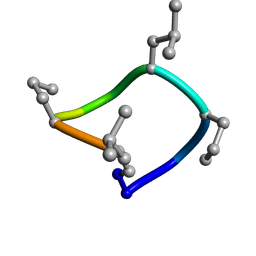 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UBE
 
 | |
7UBI
 
 | |
