3R0X
 
 | | Crystal structure of Selenomethionine incorporated apo D-serine deaminase from Salmonella tyhimurium | | Descriptor: | 1,2-ETHANEDIOL, D-serine dehydratase, SODIUM ION, ... | | Authors: | Bharath, S.R, Shveta, B, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Febs J., 2011
|
|
3R0Z
 
 | | Crystal structure of apo D-serine deaminase from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, D-serine dehydratase, SULFATE ION | | Authors: | Bharath, S.R, Shveta, B, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Febs J., 2011
|
|
4D92
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase soaked with beta-chloro-D-alanine shows pyruvate bound 4 A away from active site | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, PYRUVIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D99
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase with L-ser bound non-covalently at the active site | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, SERINE, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D96
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with 1-amino-1-carboxycyclopropane (ACC) | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9E
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with L-cycloserine (LCS) | | Descriptor: | BENZAMIDINE, D-Cysteine desulfhydrase, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9B
 
 | | Pyridoxamine 5' phosphate (PMP) bound form of Salmonella typhimurium D-Cysteine desulfhydrase obtained after co-crystallization with D-cycloserine | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9C
 
 | | PMP bound form of Salmonella typhimurium D-Cysteine desulfhydrase obtained after co-crystallization with L-cycloserine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, D-Cysteine desulfhydrase | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D8T
 
 | | Crystal structure of D-Cysteine desulfhydrase from Salmonella typhimurium at 2.2 A resolution | | Descriptor: | BENZAMIDINE, CHLORIDE ION, D-cysteine desulfhydrase, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9F
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with D-cycloserine (DCS) | | Descriptor: | BENZAMIDINE, D-Cysteine desulfhydrase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D97
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase with D-ser bound at active site | | Descriptor: | BENZAMIDINE, D-SERINE, D-cysteine desulfhydrase | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D8W
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase soaked with D-cys shows pyruvate bound 4 A away from active site | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D8U
 
 | | Crystal structure of D-Cysteine desulfhydrase from Salmonella typhimurium at 3.3 A in monoclinic space group with 8 subunits in the asymmetric unit | | Descriptor: | D-cysteine desulfhydrase, PHOSPHATE ION | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
5H3U
 
 | | Sm RNA bound to GEMIN5-WD | | Descriptor: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*AP*AP*UP*UP*UP*UP*UP*GP*AP*C)-3') | | Authors: | Bharath, S.R, Tang, X, Song, H. | | Deposit date: | 2016-10-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
5H3T
 
 | | m7G cap bound to GEMIN5-WD | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Gem-associated protein 5 | | Authors: | Bharath, S.R, Tang, X, Song, H. | | Deposit date: | 2016-10-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.571 Å) | | Cite: | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
5H3S
 
 | | apo form of GEMIN5-WD | | Descriptor: | GLYCEROL, Gem-associated protein 5 | | Authors: | Bharath, S.R, Tang, X, Song, H. | | Deposit date: | 2016-10-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
5XX1
 
 | | Crystal structure of Arginine decarboxylase (AdiA) from Salmonella typhimurium | | Descriptor: | Arginine decarboxylase, PHOSPHATE ION | | Authors: | Deka, G, Bharath, S.R, Shavithri, H.S, Murthy, M.R.N. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies on the decameric S. typhimurium arginine decarboxylase (ADC): Pyridoxal 5'-phosphate binding induces conformational changes
Biochem. Biophys. Res. Commun., 490, 2017
|
|
8SEI
 
 | | SF Tau from Down Syndrome | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hoq, M.R, Bharath, S.R, Jiang, W, Vago, F.S, Bharath, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta and tau filaments in Down syndrome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5JXE
 
 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
3USU
 
 | | Crystal structure of Butea monosperma seed lectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abhilash, J, Geethanandan, K, Bharath, S.R, Sadasivan, C, Haridas, M. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of Butea monosperma seed lectin
To be Published
|
|
6LG2
 
 | | VanR bound to Vanillate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Predicted transcriptional regulators | | Authors: | He, Y, Bharath, S.R, Song, H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps.
Nat Commun, 11, 2020
|
|
6L3G
 
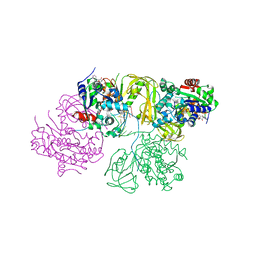 | | Structural Basis for DNA Unwinding at Forked dsDNA by two coordinating Pif1 helicases | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*TP*TP*TP*T)-3'), ... | | Authors: | Su, N, Bharath, S.R, Song, H. | | Deposit date: | 2019-10-10 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for DNA unwinding at forked dsDNA by two coordinating Pif1 helicases.
Nat Commun, 10, 2019
|
|
9CZP
 
 | |
9CZL
 
 | |
9CZI
 
 | |
