1KBL
 
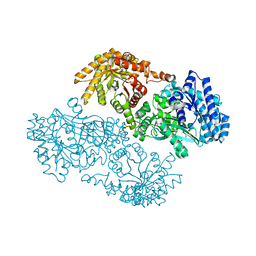 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | AMMONIUM ION, PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C, Liu, S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
1BLP
 
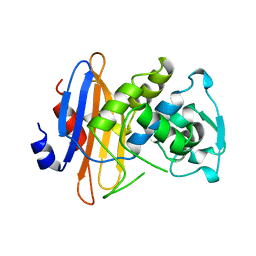 | |
3BLM
 
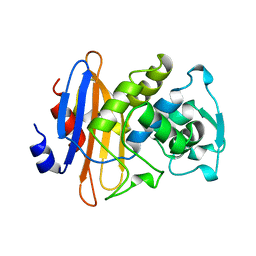 | |
1DIK
 
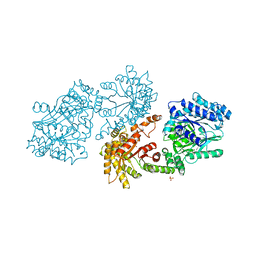 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5TNC
 
 | |
1GPR
 
 | |
1KC7
 
 | | Pyruvate Phosphate Dikinase with Bound Mg-phosphonopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHONOPYRUVATE, SULFATE ION, ... | | Authors: | Chen, C.C, Howard, A, Herzberg, O. | | Deposit date: | 2001-11-07 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
2HPR
 
 | |
7LYZ
 
 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | Deposit date: | 1977-05-06 | | Release date: | 1977-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
2DUA
 
 | |
4QT8
 
 | | Crystal Structure of RON Sema-PSI-IPT1 extracellular domains in complex with MSP beta-chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor-like protein, Macrophage-stimulating protein receptor, ... | | Authors: | Herzberg, O, Chao, K.L. | | Deposit date: | 2014-07-07 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the binding specificity of human Recepteur d'Origine Nantais (RON) receptor tyrosine kinase to macrophage-stimulating protein.
J.Biol.Chem., 289, 2014
|
|
1BSG
 
 | |
3LYE
 
 | | Crystal structure of oxaloacetate acetylhydrolase | | Descriptor: | CALCIUM ION, Oxaloacetate acetyl hydrolase | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-02-26 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
3OHI
 
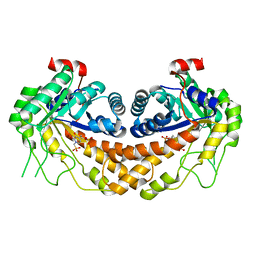 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | Descriptor: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Herzberg, O, Galkin, A. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
3M0K
 
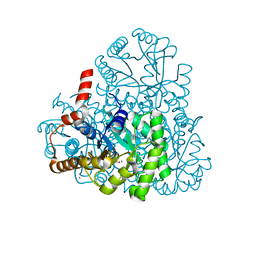 | |
3M0J
 
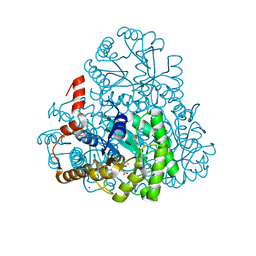 | | Structure of oxaloacetate acetylhydrolase in complex with the inhibitor 3,3-difluorooxalacetate | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-03-03 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
3QYM
 
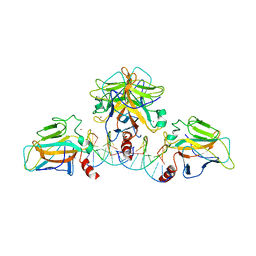 | |
1EGO
 
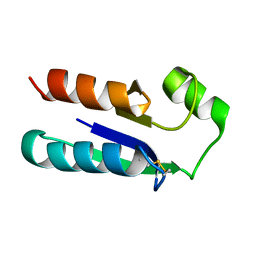 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | Descriptor: | GLUTAREDOXIN | | Authors: | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1MOU
 
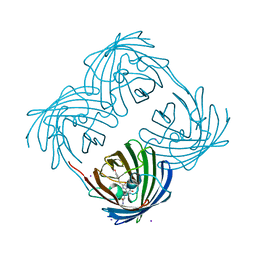 | | Crystal structure of Coral pigment | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
1MOV
 
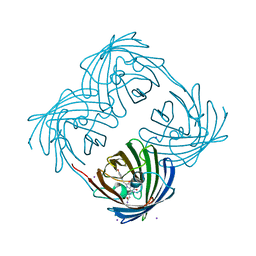 | | Crystal structure of Coral protein mutant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
2XD5
 
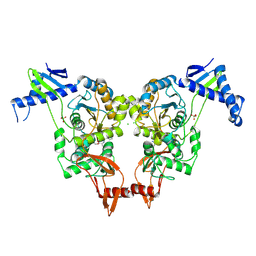 | | Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b | | Descriptor: | CHLORIDE ION, N-BENZOYL-D-ALANINE, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Lemaire, D, Jamin, M, Dideberg, O, Dessen, A. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism and the Role of Streptococcus Pneumoniae Pbp1B
To be Published
|
|
4UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
1VDR
 
 | | DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, PHOSPHATE ION | | Authors: | Pieper, U, Herzberg, O. | | Deposit date: | 1997-11-30 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii.
Structure, 6, 1998
|
|
1BLH
 
 | |
1M1B
 
 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | Authors: | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2002-06-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
