4JPB
 
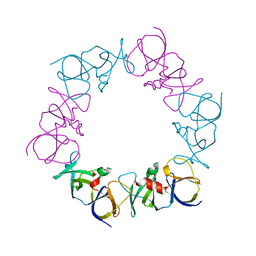 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
7XMC
 
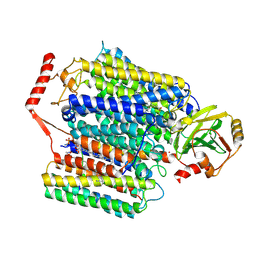 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMD
 
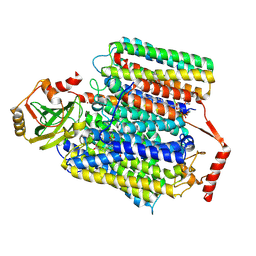 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
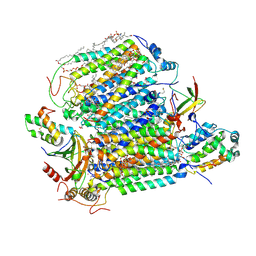 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
3WRV
 
 | |
3WRW
 
 | |
8HRX
 
 | | Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex | | Descriptor: | Fab heavy chain from antibody IgG clone number YN9048, Fab light chain from antibody IgG clone number YN9048, PreS1 protein (Fragment), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HRY
 
 | | Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex | | Descriptor: | Fab heavy chain from antibody IgG clone number YN9016, Fab light chain from antibody IgG clone number YN9016, Large S protein (Fragment), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3WRY
 
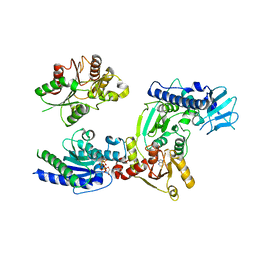 | | Crystal structure of helicase complex 2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WRX
 
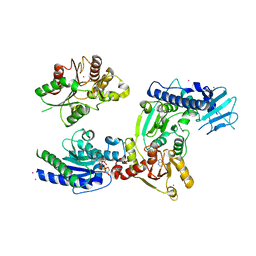 | | Crystal structure of helicase complex 1 | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7W7V
 
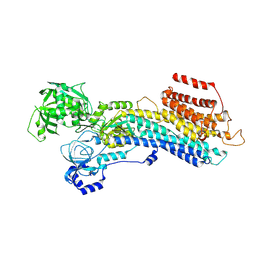 | | 'late' E2P of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7U
 
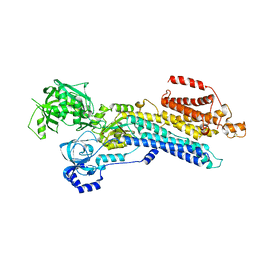 | | The 'Ca2+-unbound' BeF3- of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7W
 
 | | E2 Pi of SERCA2b | | Descriptor: | MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2, TETRAFLUOROALUMINATE ION | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7T
 
 | | The E1-BeF3- 2Ca2+ of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
