3SYN
 
 | | Crystal structure of FlhF in complex with its activator | | Descriptor: | ALUMINUM FLUORIDE, ATP-binding protein YlxH, Flagellar biosynthesis protein flhF, ... | | Authors: | Bange, G, Kuemmerer, N, Wild, K, Sinning, I. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Structural basis for the molecular evolution of SRP-GTPase activation by protein.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3MIX
 
 | | Crystal structure of the cytosolic domain of B. subtilis FlhA | | Descriptor: | Flagellar biosynthesis protein flhA | | Authors: | Bange, G, Kuemmerer, N, Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PX3
 
 | | Crystal structure of FlhF complexed with GTP/Mg(2+) | | Descriptor: | Flagellar biosynthesis protein flhF, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PX0
 
 | | Crystal structure of FlhF complexed with GMPPNP/Mg(2+) | | Descriptor: | Flagellar biosynthesis protein flhF, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4IPA
 
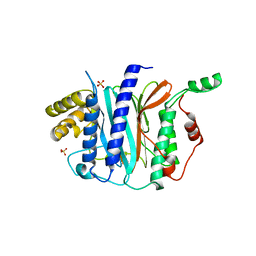 | | Structure of a thermophilic Arx1 | | Descriptor: | Putative curved DNA-binding protein, SULFATE ION | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2013-01-09 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
4GMO
 
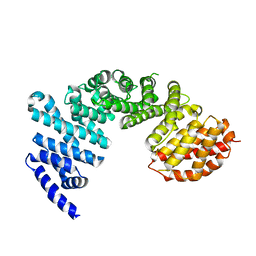 | | Crystal structure of Syo1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMN
 
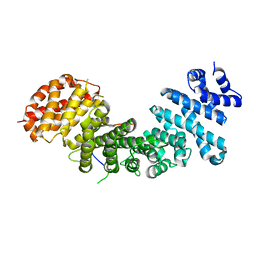 | | Structural basis of Rpl5 recognition by Syo1 | | Descriptor: | 60S ribosomal protein l5-like protein, Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GNI
 
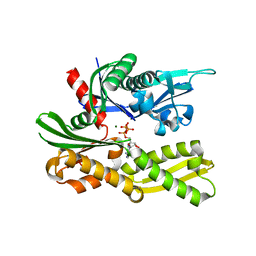 | | Structure of the Ssz1 ATPase bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6ZCW
 
 | |
6ZCV
 
 | |
5L3W
 
 | | Structure of the crenarchaeal FtsY GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Signal recognition particle receptor FtsY | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3R
 
 | | Structure of the GTPase heterodimer of chloroplast SRP54 and FtsY from Arabidopsis thaliana | | Descriptor: | Cell division protein FtsY homolog, chloroplastic, GLYCEROL, ... | | Authors: | Bange, G, Kribelbauer, J, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3V
 
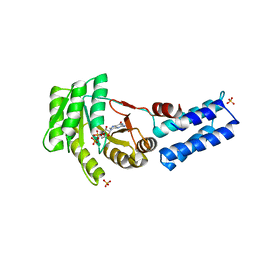 | | Structure of the crenarchaeal SRP54 GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Signal recognition particle 54 kDa protein | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3S
 
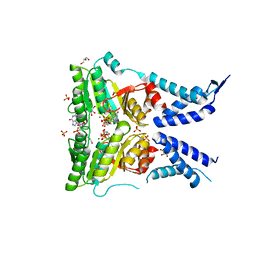 | | Structure of the GTPase heterodimer of crenarchaeal SRP54 and FtsY | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
6FGJ
 
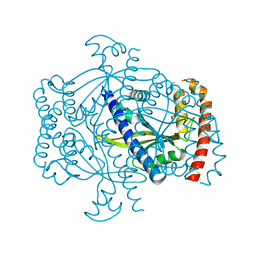 | | Crystal structure of the small alarmone synthethase 2 from Staphylococcus aureus | | Descriptor: | GTP pyrophosphokinase, TRIETHYLENE GLYCOL | | Authors: | Bange, G, Steinchen, W, Vogt, M, Altegoer, F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ.
Sci Rep, 8, 2018
|
|
6FGX
 
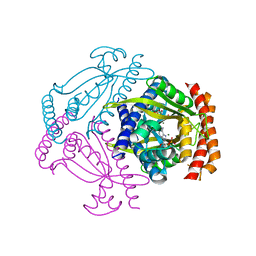 | | Crystal structure of the small alarmone synthethase 2 from Staphylococcus aureus bound to AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GTP pyrophosphokinase, MAGNESIUM ION | | Authors: | Bange, G, Vogt, M, Steinchen, W, Altegoer, F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ.
Sci Rep, 8, 2018
|
|
6FGK
 
 | |
7QCW
 
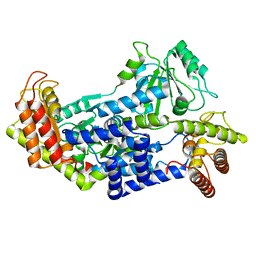 | |
6YVC
 
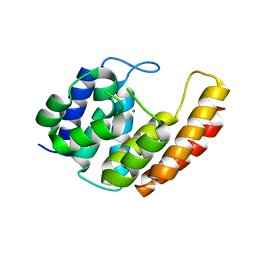 | |
5F2V
 
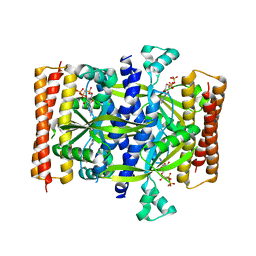 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GTP pyrophosphokinase YjbM, MAGNESIUM ION | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2YHS
 
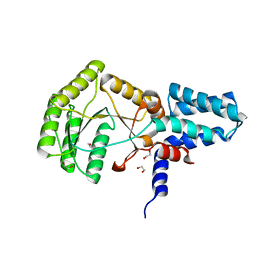 | | Structure of the E. coli SRP receptor FtsY | | Descriptor: | 1,2-ETHANEDIOL, CELL DIVISION PROTEIN FTSY | | Authors: | Stjepanovic, G, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lipids Trigger a Conformational Switch that Regulates Signal Recognition Particle (Srp)-Mediated Protein Targeting.
J.Biol.Chem., 286, 2011
|
|
7OJ1
 
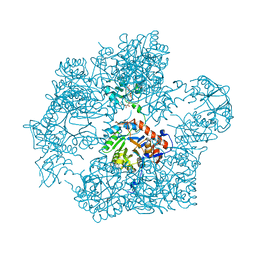 | | Bacillus subtilis IMPDH in complex with Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Giammarinaro, P.I, Bange, G. | | Deposit date: | 2021-05-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Nat Microbiol, 7, 2022
|
|
8A4O
 
 | |
6GOW
 
 | |
6H3P
 
 | |
