1E61
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
4QYZ
 
 | | Crystal structure of a CRISPR RNA-guided surveillance complex, Cascade, bound to a ssDNA target | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Mulepati, S, Bailey, S. | | Deposit date: | 2014-07-26 | | Release date: | 2014-09-03 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (3.0303 Å) | | Cite: | Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target.
Science, 345, 2014
|
|
4DMR
 
 | | REDUCED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS WITH BOUND DMSO SUBSTRATE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, DMSO REDUCTASE, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-30 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The high resolution crystal structure of DMSO reductase in complex with DMSO.
J.Mol.Biol., 275, 1998
|
|
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
3DMR
 
 | | STRUCTURE OF DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS AT PH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM(VI) ION, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-25 | | Release date: | 1998-03-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molybdenum Active Centre of Dmso Reductase from Rhodobacter Capsulatus: Crystal Structure of the Oxidised Enzyme at 1.82-A Resolution and the Dithionite-Reduced Enzyme at 2.8-A Resolution
J.Biol.Inorg.Chem., 2, 1997
|
|
2DMR
 
 | | DITHIONITE REDUCED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM(IV) ION, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-24 | | Release date: | 1998-03-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molybdenum Active Centre of Dmso Reductase from Rhodobacter Capsulatus: Crystal Structure of the Oxidised Enzyme at 1.82-A Resolution and the Dithionite-Reduced Enzyme at 2.8-A Resolution
J.Biol.Inorg.Chem., 2, 1997
|
|
5H9E
 
 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target (32-nt spacer) at 3.20 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
5H9F
 
 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
3GNA
 
 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*AP*AP*CP*AP*AP*AP*AP*AP*CP*C)-3', 5'-D(*TP*GP*GP*TP*TP*TP*TP*TP*GP*TP*TP*AP*AP*G)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GNB
 
 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*TP*TP*CP*AP*GP*AP*AP*AP*CP*C)-3', 5'-D(*AP*GP*GP*TP*TP*TP*CP*TP*GP*AP*AP*AP*AP*C)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1GNL
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans X-ray structure at 1.25A resolution using synchrotron radiation at a wavelength of 0.933A | | Descriptor: | ACETATE ION, HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GN9
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-04 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1H76
 
 | | The crystal structure of diferric porcine serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-03 | | Release date: | 2002-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal and Molecular Structures of Diferric Porcine and Rabbit Serum Transferrins at Resolutions of 2.15 And 2.60A, Respectively
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JNF
 
 | | Rabbit serum transferrin at 2.6 A resolution. | | Descriptor: | CARBONATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-24 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3CB4
 
 | | The Crystal Structure of LepA | | Descriptor: | GTP-binding protein lepA | | Authors: | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3E0D
 
 | | Insights into the Replisome from the Crystral Structure of the Ternary Complex of the Eubacterial DNA Polymerase III alpha-subunit | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase III subunit alpha, ... | | Authors: | Wing, R.A, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Insights into the Replisome from the Structure of a Ternary Complex of the DNA Polymerase III alpha-Subunit.
J.Mol.Biol., 382, 2008
|
|
3E3J
 
 | | Crystal Structure of an Intermediate Complex of T7 RNAP and 8nt of RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*DTP*DAP*DAP*DTP*DAP*DCP*DGP*DAP*DCP*DTP*DCP*DAP*DCP*DTP*DAP*DTP*DAP*DTP*DTP*DTP*DCP*DTP*DGP*DCP*DCP*DAP*DAP*DAP*DCP*DGP*DGP*DC)-3'), DNA-directed RNA polymerase, ... | | Authors: | Durniak, K.J, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-08-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Science, 322, 2008
|
|
3E2E
 
 | | Crystal Structure of an Intermediate Complex of T7 RNAP and 7nt of RNA | | Descriptor: | DNA (28-MER), DNA (31-MER), DNA-directed RNA polymerase, ... | | Authors: | Durniak, K.J, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Science, 322, 2008
|
|
1E1D
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris | | Descriptor: | HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Arendsen, A.F, Cooper, S.J, Bailey, S, Garner, C.D, Hagen, W.R, Lindley, P.F. | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-21 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Hybrid-Cluster Protein (Hcp) from Desulfovibrio Vulgaris (Hildenborough) at 1.6 A Resolution.
Biochemistry, 39, 2000
|
|
1E2U
 
 | | Low Temperature Structure of Hybrid Cluster Protein from Desulfovibrio vulgaris to 1.6A | | Descriptor: | HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Cooper, S.J, Bailey, S, Hagen, W.R, Lindley, P.F. | | Deposit date: | 2000-05-26 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hybrid-Cluster Protein (Hcp) from Desulfovibrio Vulgaris (Hildenborough) at 1.6 A Resolution.
Biochemistry, 39, 2000
|
|
1DMR
 
 | | OXIDIZED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM(VI) ION, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molybdenum Active Centre of Dmso Reductase from Rhodobacter Capsulatus: Crystal Structure of the Oxidised Enzyme at 1.82-A Resolution and the Dithionite-Reduced Enzyme at 2.8-A Resolution
J.Biol.Inorg.Chem., 2, 1997
|
|
1E9V
 
 | | XENON BOUND IN HYDROPHOBIC CHANNEL OF HYBRID CLUSTER PROTEIN FROM DESULFOVIBRIO VULGARIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Cooper, S.J, Bailey, S, Rizkallah, P.J, Lindley, P.F. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-25 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ferricyanide Soaked Hybrid Cluster Protein at 1.2A and Xenon Mapping of the Hydrophobic Cavity at 1.8A
To be Published
|
|
1AOG
 
 | |
5HG9
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | Descriptor: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG8
 
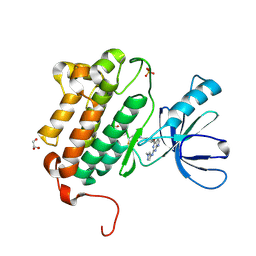 | | EGFR (L858R, T790M, V948R) in complex with N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]propanamide, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
