3T4B
 
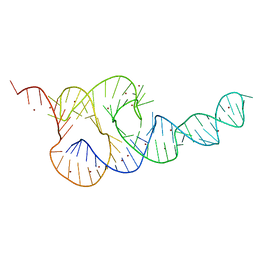 | | Crystal Structure of the HCV IRES pseudoknot domain | | 分子名称: | HCV IRES pseudoknot domain plus crystallization module, NICKEL (II) ION | | 著者 | Berry, K.E, Waghray, S, Mortimer, S.A, Bai, Y, Doudna, J.A. | | 登録日 | 2011-07-25 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning.
Structure, 19, 2011
|
|
6OGE
 
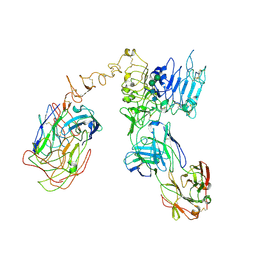 | | Cryo-EM structure of Her2 extracellular domain-Trastuzumab Fab-Pertuzumab Fab complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab FAB HEAVY CHAIN, Pertuzumab FAB LIGHT CHAIN, ... | | 著者 | Hao, Y, Yu, X, Bai, Y, Huang, X. | | 登録日 | 2019-04-02 | | 公開日 | 2019-05-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM Structure of HER2-trastuzumab-pertuzumab complex.
Plos One, 14, 2019
|
|
6BUZ
 
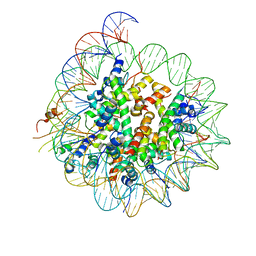 | | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B, ... | | 著者 | Chittori, S, Hong, J, Kelly, A.E, Bai, Y, Subramaniam, S. | | 登録日 | 2017-12-11 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N.
Science, 359, 2018
|
|
7K63
 
 | |
7K61
 
 | |
7K5X
 
 | |
7K5Y
 
 | |
7K60
 
 | |
7D1O
 
 | | Crystal structure of SARS-Cov-2 main protease with narlaprevir | | 分子名称: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Fu, L.F, Feng, Y, Qi, J.X. | | 登録日 | 2020-09-15 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural basis for the inhibition of the SARS-CoV-2 main protease by the anti-HCV drug narlaprevir.
Signal Transduct Target Ther, 6, 2021
|
|
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | 分子名称: | CITRIC ACID, DNA (167-mer), H5, ... | | 著者 | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | 登録日 | 2014-06-11 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.503 Å) | | 主引用文献 | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
4R0T
 
 | | Crystal structure of P. aeruginosa TpbA (C132S) in complex with pTyr | | 分子名称: | PHOSPHATE ION, Protein tyrosine phosphatase TpbA, TYROSINE | | 著者 | Xu, K, Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2014-08-01 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
Plos One, 10
|
|
4R0S
 
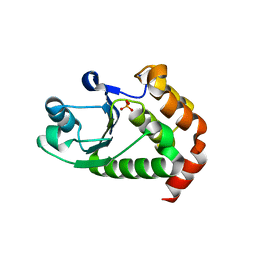 | | Crystal structure of P. aeruginosa TpbA | | 分子名称: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | 著者 | Xu, K, Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2014-08-01 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
4WYU
 
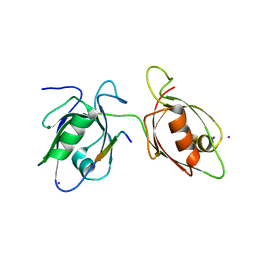 | |
4WYT
 
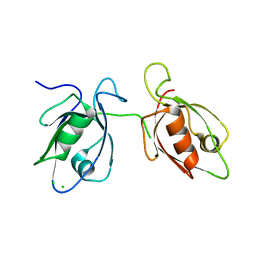 | |
4X23
 
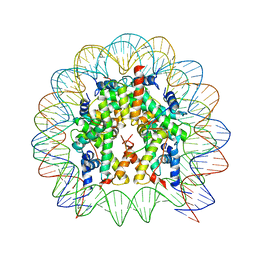 | |
7CHF
 
 | |
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | 著者 | Xiao, J, Zhu, Q, Wang, G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-11-25 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHE
 
 | |
7CH4
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | 著者 | Xiao, J, Zhu, Q. | | 登録日 | 2020-07-05 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHC
 
 | |
7CH5
 
 | |
9BHA
 
 | |
9BH8
 
 | |
9BH7
 
 | |
