1DF1
 
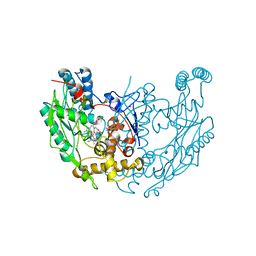 | | MURINE INOSOXY DIMER WITH ISOTHIOUREA BOUND IN THE ACTIVE SITE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Rosenfeld, R.J, Arvai, A.S, Ghosh, D.K, Ghosh, S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-16 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
EMBO J., 18, 1999
|
|
4EEU
 
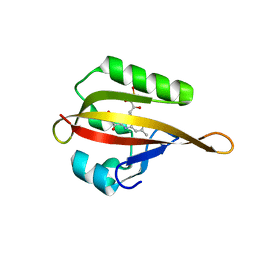 | | Crystal structure of phiLOV2.1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4068 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EER
 
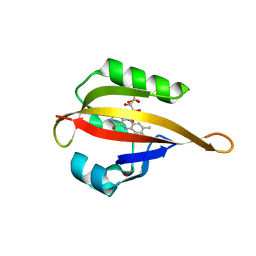 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EES
 
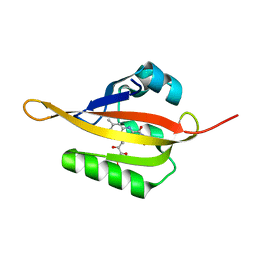 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EET
 
 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EEP
 
 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
7K07
 
 | |
7K06
 
 | |
7N8W
 
 | |
7N8V
 
 | |
1JWJ
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457F Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
1JWK
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457A Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
1KEA
 
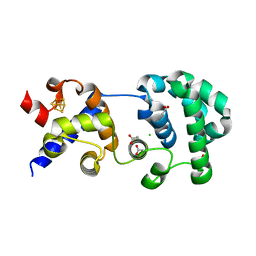 | | STRUCTURE OF A THERMOSTABLE THYMINE-DNA GLYCOSYLASE | | Descriptor: | ACETATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Mol, C.D, Arvai, A.S, Begley, T.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 2001-11-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases.
J.Mol.Biol., 315, 2002
|
|
1MRP
 
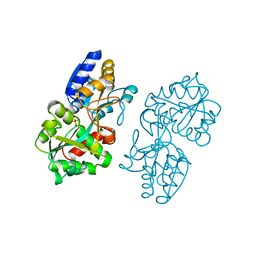 | | FERRIC-BINDING PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | FE (III) ION, FERRIC IRON BINDING PROTEIN, PHOSPHATE ION | | Authors: | Bruns, C.M, Nowalk, A.J, Arvai, A.S, Mctigue, M.A, Vaughan, K.G, Mietzner, T.A, Mcree, D.E. | | Deposit date: | 1997-05-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily.
Nat.Struct.Biol., 4, 1997
|
|
1NIW
 
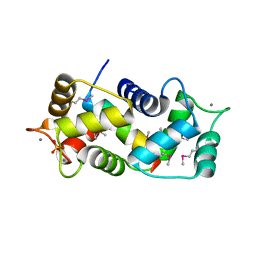 | | Crystal structure of endothelial nitric oxide synthase peptide bound to calmodulin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nitric-oxide synthase, ... | | Authors: | Aoyagi, M, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-12-26 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for endothelial nitric oxide synthase binding to calmodulin
Embo J., 22, 2003
|
|
1NOS
 
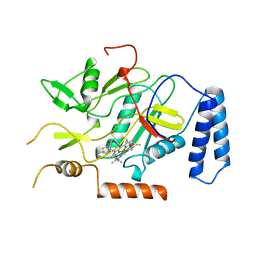 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DOMAIN (DELTA 114), IMIDAZOLE COMPLEX | | Descriptor: | IMIDAZOLE, INDUCIBLE NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1997-09-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of nitric oxide synthase oxygenase domain and inhibitor complexes.
Science, 278, 1997
|
|
1NOC
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DOMAIN (DELTA 114) COMPLEXED WITH TYPE I E. COLI CHLORAMPHENICOL ACETYL TRANSFERASE AND IMIDAZOLE | | Descriptor: | IMIDAZOLE, INDUCIBLE NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1997-09-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of nitric oxide synthase oxygenase domain and inhibitor complexes.
Science, 278, 1997
|
|
1OQW
 
 | |
3RCZ
 
 | | Rad60 SLD2 Ubc9 Complex | | Descriptor: | DNA repair protein rad60, SUMO-conjugating enzyme ubc9 | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
3RD2
 
 | | NIP45 SUMO-like Domain 2 | | Descriptor: | NFATC2-interacting protein | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
3SOJ
 
 | | Francisella tularensis pilin PilE | | Descriptor: | PilE, SULFATE ION | | Authors: | Wood, T, Arvai, A.S, Shin, D.S, Hartung, S, Kolappan, S, Craig, L, Tainer, J.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-11-02 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential.
J.Biol.Chem., 286, 2011
|
|
4O24
 
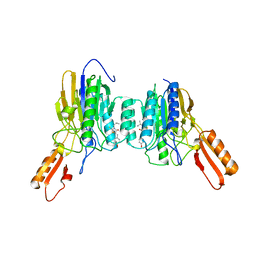 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{Z})-5-[(4-hydroxyphenyl)methylidene]-3-(2-methylpropyl)-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4NZV
 
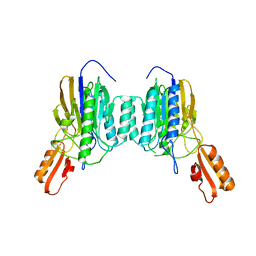 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | Exonuclease, putative, MANGANESE (II) ION | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4O4K
 
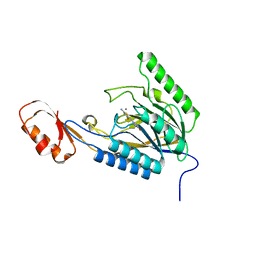 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-2-azanylidene-5-[(4-hydroxyphenyl)methylidene]-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4O43
 
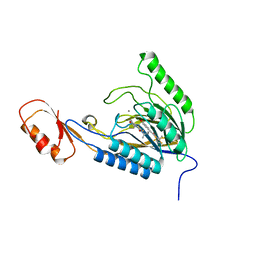 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-3-[(2~{R})-butan-2-yl]-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
