8AU0
 
 | |
8B46
 
 | | Crystal structure of the SUN1-KASH6 9:9 complex | | Descriptor: | CHLORIDE ION, Inositol 1,4,5-triphosphate receptor associated 2, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Erlandsen, B.S, Davies, O.R. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Commun Biol, 7, 2024
|
|
7Z8Z
 
 | |
7Z8Y
 
 | | Crystal structure of the SUN1-KASH6 9:6 complex | | Descriptor: | CHLORIDE ION, Inositol 1,4,5-triphosphate receptor associated 2, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2022-03-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Commun Biol, 7, 2024
|
|
6R16
 
 | | Crystal structure of the SUN1-KASH4 6:6 complex | | Descriptor: | 1,2-ETHANEDIOL, Nesprin-4, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife, 10, 2021
|
|
6R2I
 
 | | Crystal structure of the SUN1-KASH5 6:6 complex | | Descriptor: | KASH5, POTASSIUM ION, SUN domain-containing protein 1 | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2019-03-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife, 10, 2021
|
|
1NU3
 
 | | Limonene-1,2-epoxide hydrolase in complex with valpromide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PROPYLPENTANAMIDE, limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
6R15
 
 | | Crystal structure of the SUN1-KASH1 6:6 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nesprin-1, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife, 10, 2021
|
|
1NWW
 
 | | Limonene-1,2-epoxide hydrolase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEPTANAMIDE, Limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
4MBT
 
 | | Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins | | Descriptor: | GLYCEROL, VVTL1 | | Authors: | Marangon, M, Menz, R.I, Waters, E.J, Van Sluyter, S.C. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Haze Forming Proteins in White Wines: Vitis vinifera Thaumatin-Like Proteins.
Plos One, 9, 2014
|
|
4L5H
 
 | | Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins | | Descriptor: | GLYCEROL, VVTL1 | | Authors: | Marangon, M, Menz, R.I, Waters, E.J, Van Sluyter, S.C. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Haze Forming Proteins in White Wines: Vitis vinifera Thaumatin-Like Proteins.
Plos One, 9, 2014
|
|
4JRU
 
 | | Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins | | Descriptor: | GLYCEROL, thaumatin-like protein | | Authors: | Marangon, M, Menz, R.I, Waters, E.J, Van Sluyter, S.C. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Haze Forming Proteins in White Wines: Vitis vinifera Thaumatin-Like Proteins.
Plos One, 9, 2014
|
|
4OA3
 
 | | Crystal structure of the BA42 protein from BIZIONIA ARGENTINENSIS | | Descriptor: | CALCIUM ION, PROTEIN BA42 | | Authors: | Otero, L.H, Klinke, S, Aran, M, Pellizza, L, Goldbaum, F.A, Cicero, D. | | Deposit date: | 2014-01-03 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
2MPB
 
 | | NMR structure of BA42 protein from the psychrophilic bacteria Bizionia argentinensis sp. nov | | Descriptor: | BA42 | | Authors: | Cicero, D, Aran, M, Smal, C, Pellizza, L, Gallo, M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
8B5X
 
 | | Crystal structure of the SUN1-KASH6 9:6 complex | | Descriptor: | CHLORIDE ION, Inositol 1,4,5-triphosphate receptor associated 2, POTASSIUM ION, ... | | Authors: | Erlandsen, B.S, Gurusaran, M, Davies, O.R. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Commun Biol, 7, 2024
|
|
3P4A
 
 | | 2'Fluoro modified RNA octamer fA2U2 | | Descriptor: | 2'Fluoro modified RNA 8-MER, MAGNESIUM ION, STRONTIUM ION | | Authors: | Manoharan, M, Akinc, A, Pandey, R.K, Qin, J, Hadwiger, P, John, M, Mills, K, Charisse, K, Maier, M.A, Nechev, L, Greene, E.M, Pallan, P.S, Rozners, E, Rajeev, K.G, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
3O21
 
 | | High resolution structure of GluA3 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3, PHOSPHATE ION | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Babu, M.M, Jensen, M.H, Greger, I.H. | | Deposit date: | 2010-07-22 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Dynamics and allosteric potential of the AMPA receptor N-terminal domain
Embo J., 30, 2011
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6GNX
 
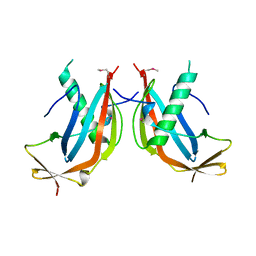 | | Crystal structure of the MAJIN-TERB2 heterotetrameric complex - selenomethionine derivative | | Descriptor: | Membrane-anchored junction protein, Telomere repeats-binding bouquet formation protein 2 | | Authors: | Gurusaran, M, Dunce, J.M, Sen, L.T, Davies, O.R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-12 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of meiotic telomere attachment to the nuclear envelope by MAJIN-TERB2-TERB1.
Nat Commun, 9, 2018
|
|
3P3W
 
 | |
4GPA
 
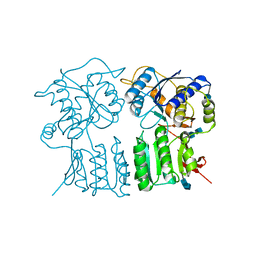 | | High resolution structure of the GluA4 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 4 | | Authors: | Sukumaran, M, Greger, I.H. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparative Dynamics of NMDA- and AMPA-Glutamate Receptor N-Terminal Domains.
Structure, 20, 2012
|
|
7TBR
 
 | |
6OV6
 
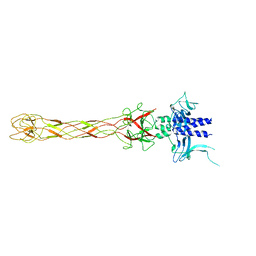 | | CRYSTALLOGRAPHIC STRUCTURE OF THE C24 PROTEIN FROM THE ANTARCTIC MICROORGANISM BIZIONIA ARGENTINENSIS | | Descriptor: | C24 PROTEIN, MANGANESE (II) ION | | Authors: | Klinke, S, Rinaldi, J, Guimaraes, B.G, Pellizza, L, Aran, M. | | Deposit date: | 2019-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the putative long tail fiber receptor-binding tip of a novel temperate bacteriophage from the Antarctic bacterium Bizionia argentinensis JUB59.
J.Struct.Biol., 212, 2020
|
|
2LT2
 
 | |
4F0Q
 
 | | MspJI Restriction Endonuclease - P21 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
