9KZ4
 
 | |
4OCP
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate and ADP | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCV
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCK
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCJ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SODIUM ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCU
 
 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SULFATE ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCO
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCQ
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4XBR
 
 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XBU
 
 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4KWE
 
 | | GDP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Li, Y, Hsin, J, Zhao, L, Cheng, Y, Shang, W, Huang, K.C, Wang, H.W, Ye, S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation
Science, 341, 2013
|
|
3RSN
 
 | | Crystal Structure of the N-terminal region of Human Ash2L | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Chen, Y, Wan, B, Wang, K.C, Cao, F, Yang, Y, Protacio, A, Dou, Y, Chang, H.Y, Lei, M. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding.
Embo Rep., 12, 2011
|
|
5ZUE
 
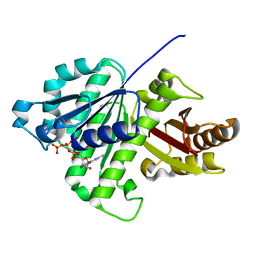 | | GTP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Guan, F, Yu, J, Yu, J, Liu, Y, Li, Y, Feng, X.H, Huang, K.C, Chang, Z, Ye, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division
Elife, 7, 2018
|
|
2JEJ
 
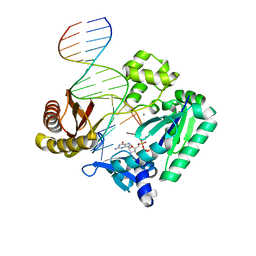 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*CP*G)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEG
 
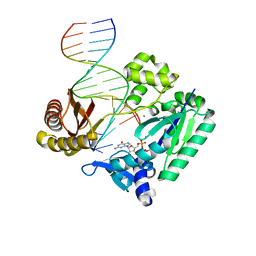 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEI
 
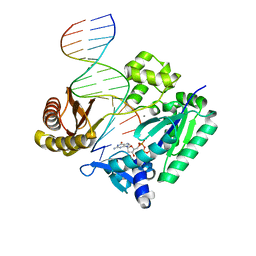 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*T)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEF
 
 | | The Molecular Basis of Selectivity of Nucleotide Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state and X-Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*CP*BZGP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JSF
 
 | |
8Z1J
 
 | | SFX structure of dark adapted CraCRY | | Descriptor: | Animal-like cryptochrome, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-11 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z24
 
 | | SFX structure of CraCRY 10 ns after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z26
 
 | | SFX structure of CraCRY 30 ns after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z2D
 
 | | SFX structure of CraCRY 100 ns after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z3G
 
 | | SFX structure of CraCRY 3 us after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-15 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z4P
 
 | | SFX structure of CraCRY 33 ms after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-17 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
8Z41
 
 | | SFX structure of CraCRY 100 us after photoexcitation of the oxidized protein | | Descriptor: | CHLORIDE ION, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-04-16 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
