8COD
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Saleem-Batcha, R, Popadic, D, Koeppl, L.H, Andexer, J.N. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine
To Be Published
|
|
8OVH
 
 | |
4L4Q
 
 | | Methionine Adenosyltransferase | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Schlesier, J, Siegrist, J, Gerhardt, S, Andexer, J.N, Einsle, O. | | Deposit date: | 2013-06-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterisation of the methionine adenosyltransferase from Thermococcus kodakarensis.
Bmc Struct.Biol., 13, 2013
|
|
8R4Z
 
 | |
5A3K
 
 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
5AG3
 
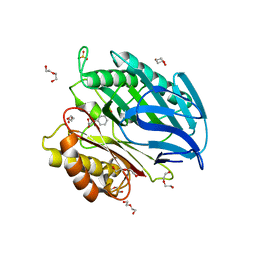 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-(2-CARBOXYETHYL)BENZOIC ACID, DI(HYDROXYETHYL)ETHER, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, ... | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-01-28 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
4BPS
 
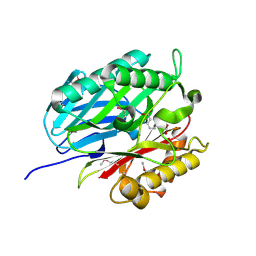 | | Crystal structure of Chorismatase at 1.08 Angstrom resolution. | | Descriptor: | 3-(2-CARBOXYETHYL)BENZOIC ACID, FKBO | | Authors: | Juneja, P, Hubrich, F, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-18 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Mechanistic Implications for the Chorismatase Fkbo Based on the Crystal Structure.
J.Mol.Biol., 426, 2014
|
|
7R39
 
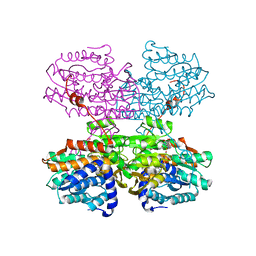 | |
7R3A
 
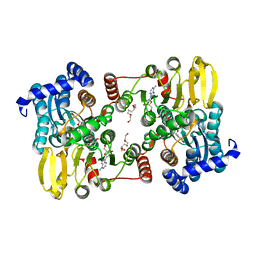 | |
7R37
 
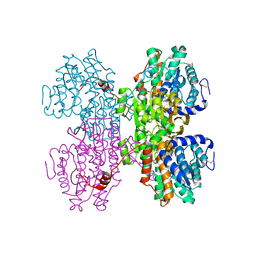 | |
7R38
 
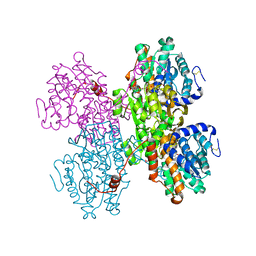 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Pyrococcus furiosus in complex with S-inosyl-L-homocysteine | | Descriptor: | (2S)-2-AMINO-4-({[(2S,3S,4R,5R)-3,4-DIHYDROXY-5-(6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)TETRAHYDROFURAN-2-YL]METHYL}THIO)BUTANOIC ACID, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Popadic, D, Andexer, J.N. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Pyrococcus furiosus in complex with S-inosyl-L-homocysteine
To Be Published
|
|
8RVC
 
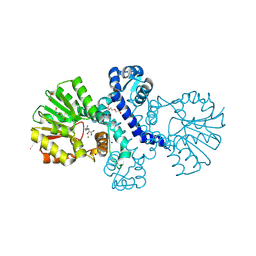 | | Crystal structure of alpha keto acid C-methyl-transferases MrsA bound to ketoarginine | | Descriptor: | 1,2-ETHANEDIOL, 2-ketoarginine methyltransferase, 5-[(diaminomethylidene)amino]-2-oxopentanoic acid, ... | | Authors: | Gerhardt, S, Kemper, F, Andexer, J.N. | | Deposit date: | 2024-02-01 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structures and protein engineering of the alpha-keto acid C-methyltransferases SgvM and MrsA for rational substrate transfer.
Chembiochem, 2024
|
|
8RWM
 
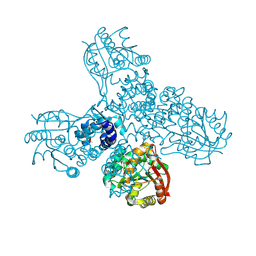 | |
8RWW
 
 | |
8RVS
 
 | |
8RXF
 
 | |
3DQZ
 
 | | Structure of the hydroxynitrile lyase from Arabidopsis thaliana | | Descriptor: | Alpha-hydroxynitrile lyase-like protein, CHLORIDE ION | | Authors: | Andexer, J, Staunig, N, Gruber, K. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Hydroxynitrile lyases with alpha / beta-hydrolase fold: two enzymes with almost identical 3D structures but opposite enantioselectivities and different reaction mechanisms
Chembiochem, 13, 2012
|
|
6Z82
 
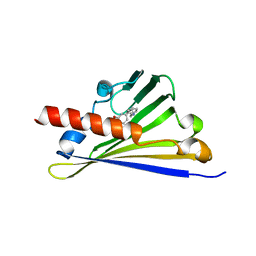 | |
5O6M
 
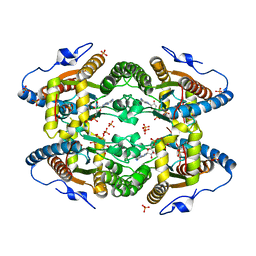 | |
5O6K
 
 | |
5LC9
 
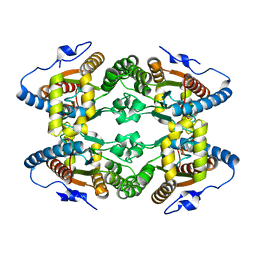 | |
5LHM
 
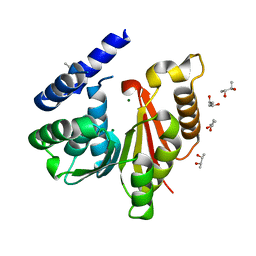 | | Crystal Structure of SafC from Myxococcus xanthus apo-Form | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Netzer, J, Einsle, O. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Functional and structural characterisation of a bacterial O-methyltransferase and factors determining regioselectivity.
FEBS Lett., 591, 2017
|
|
5LLF
 
 | |
5LOG
 
 | | Crystal Structure of SafC from Myxococcus xanthus bound to SAM | | Descriptor: | CHLORIDE ION, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Netzer, J, Einsle, O. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and structural characterisation of a bacterial O-methyltransferase and factors determining regioselectivity.
FEBS Lett., 591, 2017
|
|
5LDB
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
