3G10
 
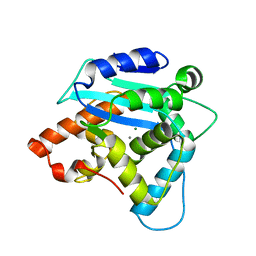 | | Structure of S. pombe Pop2p - Mg2+ and Mn2+ bound form | | 分子名称: | CCR4-Not complex subunit Caf1, MAGNESIUM ION, MANGANESE (II) ION | | 著者 | Andersen, K.R, Jonstrup, A.T, Van, L.B, Brodersen, D.E. | | 登録日 | 2009-01-29 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site
Rna, 15, 2009
|
|
3G0Z
 
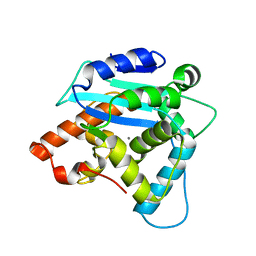 | | Structure of S. pombe Pop2p - Zn2+ and Mn2+ bound form | | 分子名称: | CCR4-Not complex subunit Caf1, MANGANESE (II) ION, ZINC ION | | 著者 | Andersen, K.R, Jonstrup, A.T, Van, L.B, Brodersen, D.E. | | 登録日 | 2009-01-29 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site
Rna, 15, 2009
|
|
5LOI
 
 | |
4KF7
 
 | |
4KF8
 
 | |
5NLU
 
 | | Structure of Nb36 crystal form 1 | | 分子名称: | SULFATE ION, single domain llama antibody Nb36 | | 著者 | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-31 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.193 Å) | | 主引用文献 | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8PEH
 
 | | Crystal structure of Lotus japonicus SYMRK kinase domain D738N | | 分子名称: | 1,2-ETHANEDIOL, Receptor-like kinase SYMRK, SULFATE ION | | 著者 | Noergaard, M.M.M, Gysel, K, Hansen, S.B, Andersen, K.R. | | 登録日 | 2023-06-14 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Phosphorylation of the alpha-I motif in SYMRK drives root nodule organogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4V7J
 
 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | 登録日 | 2009-11-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
4V7K
 
 | | Structure of RelE nuclease bound to the 70S ribosome (postcleavage state) | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | 登録日 | 2009-11-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
6EHG
 
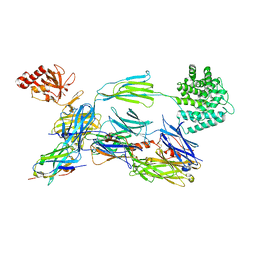 | | complement component C3b in complex with a nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Jensen, R.K, Andersen, K.R, Gadeberg, T.A.F, Laursen, N.S, Andersen, G.R. | | 登録日 | 2017-09-13 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement.
J. Biol. Chem., 293, 2018
|
|
3PRX
 
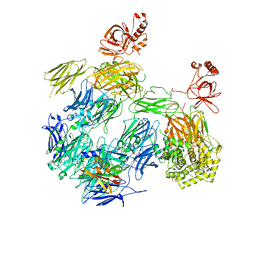 | | Structure of Complement C5 in Complex with CVF and SSL7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | 著者 | Laursen, N.S, Andersen, G.R, Sottrup-Jensen, L, Andersen, K.R, Spillner, E, Braren, I. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Embo J., 30, 2011
|
|
3PVM
 
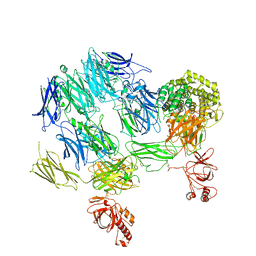 | | Structure of Complement C5 in Complex with CVF | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, Complement C5 | | 著者 | Laursen, N.S, Andersen, K.R, Braren, I, Sottrup-Jensen, L, Spillner, E, Andersen, G.R. | | 登録日 | 2010-12-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Embo J., 30, 2011
|
|
5NLW
 
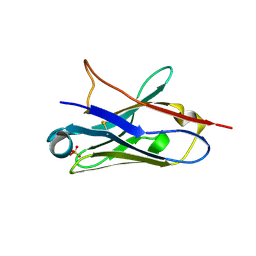 | | Structure of Nb36 crystal form 2 | | 分子名称: | SULFATE ION, nanobody Nb36 | | 著者 | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-31 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NM0
 
 | | Nb36 Ser85Cys with Hg, crystal form 1 | | 分子名称: | MERCURY (II) ION, Nb36 | | 著者 | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | 登録日 | 2017-04-05 | | 公開日 | 2017-06-07 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NML
 
 | | Nb36 Ser85Cys with Hg bound | | 分子名称: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | 著者 | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | 登録日 | 2017-04-06 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7AU7
 
 | | Crystal structure of Nod Factor Perception ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | 著者 | Gysel, K, Blaise, M, Andersen, K.R. | | 登録日 | 2020-11-02 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.547 Å) | | 主引用文献 | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BAX
 
 | | Crystal structure of LYS11 ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase | | 著者 | Laursen, M, Cheng, J, Gysel, K, Blaise, M, Andersen, K.R. | | 登録日 | 2020-12-16 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XWE
 
 | | Crystal structure of LYK3 ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | 著者 | Gysel, K, Blaise, M, Andersen, K.R. | | 登録日 | 2020-01-23 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
6QUP
 
 | | Structural signatures in EPR3 define a unique class of plant carbohydrate receptors | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, ... | | 著者 | Wong, J.E, Gysel, K, Birkefeldt, T.G, Vinther, M, Muszynski, A, Azadi, P, Laursen, N.S, Sullivan, J.T, Ronson, C.W, Stougaard, J, Andersen, K.R. | | 登録日 | 2019-02-28 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Structural signatures in EPR3 define a unique class of plant carbohydrate receptors.
Nat Commun, 11, 2020
|
|
5LS2
 
 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | 著者 | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | 登録日 | 2016-08-22 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2P51
 
 | | Crystal structure of the S. pombe Pop2p deadenylation subunit | | 分子名称: | MAGNESIUM ION, SPCC18.06c protein | | 著者 | Thyssen Jonstrup, A, Andersen, K.R, Van, L.B, Brodersen, D.E. | | 登録日 | 2007-03-14 | | 公開日 | 2007-05-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme
Nucleic Acids Res., 35, 2007
|
|
6X07
 
 | |
4FXI
 
 | |
6X08
 
 | |
6X06
 
 | |
