1A78
 
 | | COMPLEX OF TOAD OVARY GALECTIN WITH THIO-DIGALACTOSE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GALECTIN-1, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Amzel, L.M, Bianchet, M.A, Ahmed, H, Vasta, G.R. | | Deposit date: | 1998-03-20 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes
Proteins, 40, 2000
|
|
1GAN
 
 | | COMPLEX OF TOAD OVARY GALECTIN WITH N-ACETYLGALACTOSE | | Descriptor: | GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Amzel, L.M, Bianchet, M.A, Ahmed, H, Vasta, G.R. | | Deposit date: | 1996-11-06 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes.
Proteins, 40, 2000
|
|
4OLO
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Clostridiales bacterium 1_7_47FAA | | Descriptor: | BMC domain protein | | Authors: | Thompson, M.C, Ahmed, H, McCarty, K.N, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
2R91
 
 | | Crystal Structure of KD(P)GA from T.tenax | | Descriptor: | 2-Keto-3-deoxy-(6-phospho-)gluconate aldolase, SULFATE ION | | Authors: | Pauluhn, A, Pohl, E, Lorentzen, E, Siebers, B, Ahmed, H, Buchinger, S, Schomburg, D. | | Deposit date: | 2007-09-12 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and stereochemical studies of KD(P)G aldolase from Thermoproteus tenax.
Proteins, 72, 2008
|
|
1A3K
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN GALECTIN-3 CARBOHYDRATE RECOGNITION DOMAIN (CRD) AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seetharaman, J, Kanigsberg, A, Slaaby, R, Leffler, H, Barondes, S.H, Rini, J.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution.
J.Biol.Chem., 273, 1998
|
|
7SWT
 
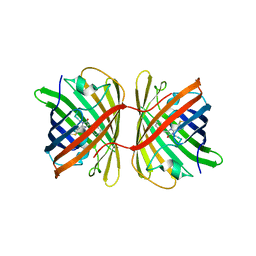 | | Crystal structure of the chromoprotein eforRED | | Descriptor: | Chromoprotein eforRED | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWS
 
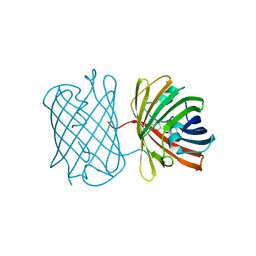 | | Crystal structure of the chromoprotein amilCP | | Descriptor: | BROMIDE ION, CHLORIDE ION, Chromoprotein amilCP | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWU
 
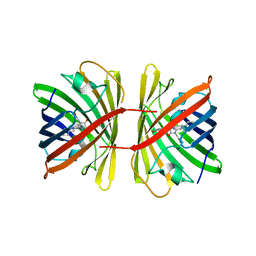 | | Crystal structure of the chromoprotein spisPINK | | Descriptor: | Chromoprotein spisPINK | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWR
 
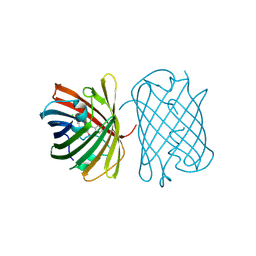 | | Crystal structure of the chromoprotein gfasPurple | | Descriptor: | CHLORIDE ION, Chromoprotein gfasPurple | | Authors: | Caputo, A.T, Newman, J, Peat, T.S, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4RCJ
 
 | | Crystal structure of YTHDF1 YTH domain in complex with 5mer m6A RNA | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
4RCM
 
 | | Crystal structure of the Pho92 YTH domain in complex with m6A | | Descriptor: | Methylated RNA-binding protein 1, RNA (5'-R(*UP*G)-D(*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
4RCI
 
 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
1SLT
 
 | |
1UXQ
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXV
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXR
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXT
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXN
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXU
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXP
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
4OLP
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Pectobacterium wasabiae | | Descriptor: | GrpU microcompartment shell protein | | Authors: | Wheatley, N.M, Thompson, M.C, Gidaniyan, S.D, Sawaya, M.R, Jorda, J, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
2R94
 
 | | Crystal Structure of KD(P)GA from T.tenax | | Descriptor: | 2-Keto-3-deoxy-(6-phospho-)gluconate aldolase, PYRUVIC ACID | | Authors: | Pauluhn, A, Pohl, E. | | Deposit date: | 2007-09-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and stereochemical studies of KD(P)G aldolase from Thermoproteus tenax.
Proteins, 72, 2008
|
|
6E20
 
 | | Crystal structure of the Dario rerio galectin-1-L2 | | Descriptor: | Galectin, MAGNESIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ghosh, A, Bianchet, M.A. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein.
Glycobiology, 29, 2019
|
|
