3GA4
 
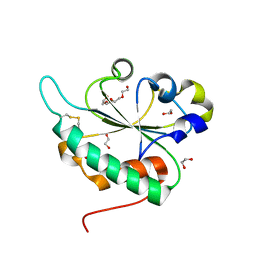 | | Crystal structure of Ost6L (photoreduced form) | | Descriptor: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G9B
 
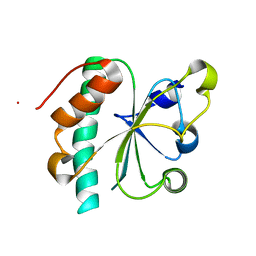 | | Crystal structure of reduced Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4M92
 
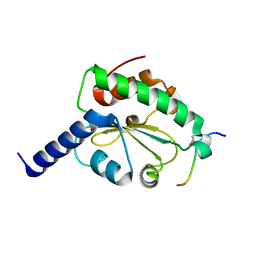 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
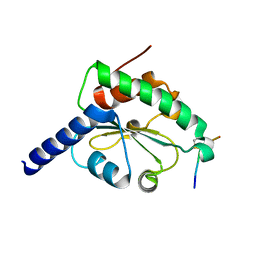 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M8G
 
 | | Crystal structure of Se-Met hN33/Tusc3 | | Descriptor: | Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-13 | | Release date: | 2014-03-26 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M90
 
 | | crystal structure of oxidized hN33/Tusc3 | | Descriptor: | Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
2MN5
 
 | | NMR structure of Copsin | | Descriptor: | Copsin | | Authors: | Hofmann, D, Wider, G, Essig, A, Aebi, M. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Copsin, a Novel Peptide-based Fungal Antibiotic Interfering with the Peptidoglycan Synthesis.
J.Biol.Chem., 289, 2014
|
|
5OGL
 
 | | Structure of bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Substrate mimicking peptide, ... | | Authors: | Napiorkowska, M, Boilevin, J, Sovdat, T, Darbre, T, Reymond, J.-L, Aebi, M, Locher, K.P. | | Deposit date: | 2017-07-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4USO
 
 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | Descriptor: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
6GEW
 
 | | OphA Y63F-sinefungin complex | | Descriptor: | OphA, S-ADENOSYL-L-HOMOCYSTEINE, SINEFUNGIN | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2018-04-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
4USP
 
 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
6ZU2
 
 | | CML1 crystal structure in complex with H-type 1 trisaccharide | | Descriptor: | Mucin-binding lectin 1, SULFATE ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Varrot, A, Bleuler-Martinez, S. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
6ZV5
 
 | | CML1 crystal structure in complex with Lewis a tetrasaccharide | | Descriptor: | Mucin-binding lectin 1, SULFATE ION, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Varrot, A, Bleuler-Martinez, S. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
5N0V
 
 | |
5N0O
 
 | |
5N0W
 
 | |
5N0Q
 
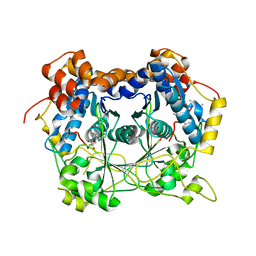 | |
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5C76
 
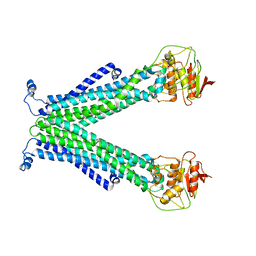 | |
5C73
 
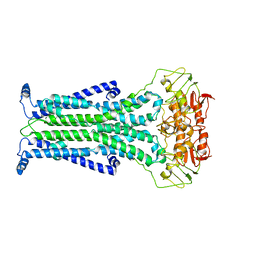 | |
5C78
 
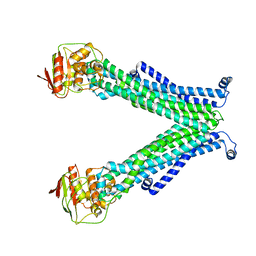 | |
5N0N
 
 | |
5N0U
 
 | |
5N0P
 
 | | Crystal structure of OphA-DeltaC18 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, peptide N-methyltranferase | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5N0X
 
 | | Crystal structure of OphA-DeltaC6 in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Peptide N-Methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
