5OA6
 
 | | Crystal structure of ScGas2 in complex with compound 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-quinolin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9O
 
 | | Crystal structure of ScGas2 in complex with compound 7. | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[4-(naphthalen-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]-3,5-bis(oxidanyl)oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Fang, W, Gomollon-Bel, F, Castro-Lopez, J, Navratilova, I, van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9Y
 
 | | Crystal structure of ScGas2 in complex with compound 11 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-pyridin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9Q
 
 | | Crystal structure of ScGas2 in complex with compound 6 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(phenylmethoxymethyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,3-beta-glucanosyltransferase GAS2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O6C
 
 | | Crystal Structure of a threonine-selective RCR E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2, ZINC ION | | Authors: | Pao, K.-C, Rafie, K.Z, van Aalten, D, Virdee, S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity.
Nature, 556, 2018
|
|
5O9P
 
 | | Crystal structure of Gas2 in complex with compound 10 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(pyridin-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5OA2
 
 | | Crystal structure of ScGas2 in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, SULFATE ION, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9R
 
 | | Crystal structure of ScGas2 in complex with compound 9 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(thiophen-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5BNW
 
 | | The active site of O-GlcNAc transferase imposes constraints on substrate sequence | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, laminB1 residues 179-191 | | Authors: | Pathak, S, Alonso, J, Schimpl, M, Rafie, K, Blair, D.E, Borodkin, V.S, Albarbarawi, O, van Aalten, D.M.F. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5C1D
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RB2L) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, PHOSPHATE ION, Retinoblastoma-like protein 2, ... | | Authors: | Schimpl, M, Rafie, K, van Aalten, D.M.F. | | Deposit date: | 2015-06-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1O6I
 
 | | Chitinase B from Serratia marcescens complexed with the catalytic intermediate mimic cyclic dipeptide CI4. | | Descriptor: | Chitinase, GLYCEROL, SULFATE ION, ... | | Authors: | Houston, D.R, Eggleston, I, Synstad, B, Eijsink, V.G.H, van Aalten, D.M.F. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The cyclic dipeptide CI-4 [cyclo-(l-Arg-d-Pro)] inhibits family 18 chitinases by structural mimicry of a reaction intermediate.
Biochem. J., 368, 2002
|
|
5DIY
 
 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
5DJS
 
 | | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M | | Descriptor: | Tetratricopeptide TPR_2 repeat protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
2J4O
 
 | | Structure of TAB1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 1 | | Authors: | van Aalten, D. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-04 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tak1-Binding Protein 1 is a Pseudophosphatase.
Biochem.J., 399, 2006
|
|
5FIH
 
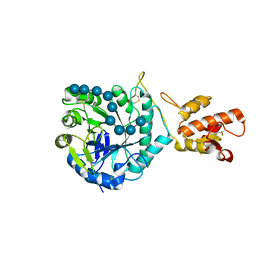 | | SACCHAROMYCES CEREVISIAE GAS2P (E176Q MUTANT) IN COMPLEX WITH LAMINARITETRAOSE AND LAMINARIPENTAOSE | | Descriptor: | 1,3-BETA-GLUCANOSYLTRANSFERASE, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Raich, L, Borodkin, V, van Aalten, D.M.F, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Trapped Covalent Intermediate of a Glycoside Hydrolase on the Pathway to Transglycosylation. Insights from Experiments and Quantum Mechanics/Molecular Mechanics Simulations.
J. Am. Chem. Soc., 138, 2016
|
|
5LVV
 
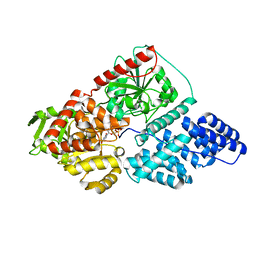 | | Human OGT in complex with UDP and fused substrate peptide (Tab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Raimi, O. | | Deposit date: | 2016-09-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
3CHE
 
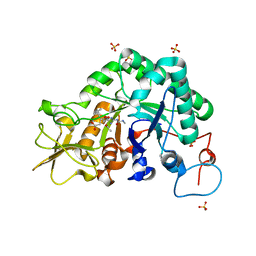 | |
3CHC
 
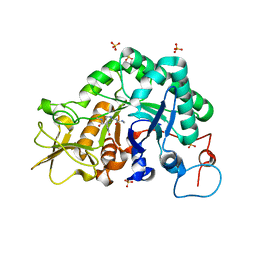 | |
3CH9
 
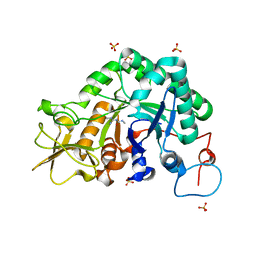 | |
3CHF
 
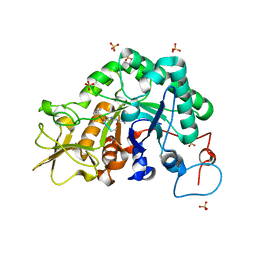 | |
3CHD
 
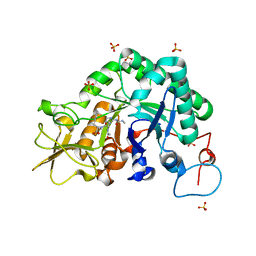 | |
5OAT
 
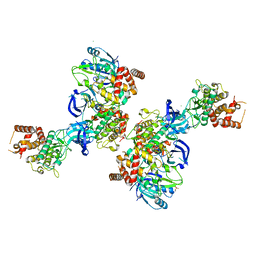 | | PINK1 structure | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein kinase PINK1, mitochondrial-like Protein | | Authors: | Kumar, A, Tamjar, J, Woodroof, H.I, Raimi, O.G, Waddell, A.Y, Peggie, M, Muqit, M.M.K, van Aalten, D.M.F. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Elife, 6, 2017
|
|
1H0I
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H0G
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys | | Descriptor: | Argadin, CHITINASE B, GLYCEROL | | Authors: | Houston, D, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2J62
 
 | | Structure of a bacterial O-glcnacase in complex with glcnacstatin | | Descriptor: | CHLORIDE ION, N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-2-METHYLPROPANAMIDE, O-GlcNAcase NagJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Shepherd, S.M, Shpiro, N.A, van Aalten, D.M.F. | | Deposit date: | 2006-09-22 | | Release date: | 2007-02-13 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels.
J. Am. Chem. Soc., 128, 2006
|
|
