4ARJ
 
 | | Crystal structure of a pesticin (translocation and receptor binding domain) from Y. pestis and T4-lysozyme chimera | | Descriptor: | PESTICIN, LYSOZYME, SULFATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-09 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARL
 
 | | Structure of the inactive pesticin D207A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARM
 
 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARP
 
 | | Structure of the inactive pesticin E178A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARQ
 
 | | Structure of the pesticin S89C, S285C double mutant | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
2XUL
 
 | | Structure of PII from Synechococcus elongatus in complex with 2- oxoglutarate at high 2-OG concentrations | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zeth, K, Chellamuthu, V.-R, Forchhammer, K, Fokina, O. | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of 2-Oxoglutarate Signaling by the Synechococcus Elongatus Pii Signal Transduction Protein
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XZW
 
 | | STRUCTURE OF PII FROM SYNECHOCOCCUS ELONGATUS IN COMPLEX WITH 2- OXOGLUTARATE AT LOW 2-OG CONCENTRATIONS | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zeth, K, Chellamuthu, V.-R, Forchhammer, K, Fokina, O. | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of 2-Oxoglutarate Signaling by the Synechococcus Elongatus Pii Signal Transduction Protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2Y21
 
 | |
2Y0T
 
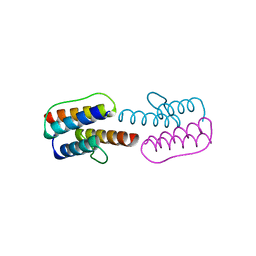 | |
2Y20
 
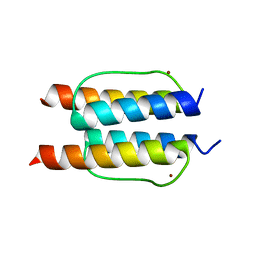 | | The mechanisms of HAMP-mediated signaling in transmembrane receptors - the A291I mutant | | Descriptor: | UNCHARACTERIZED PROTEIN, ZINC ION | | Authors: | Zeth, K, Ferris, H.U, Hulko, M, Lupas, A.N. | | Deposit date: | 2010-12-12 | | Release date: | 2011-07-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanisms of Hamp-Mediated Signaling in Transmembrane Receptors.
Structure, 19, 2011
|
|
2Y0Q
 
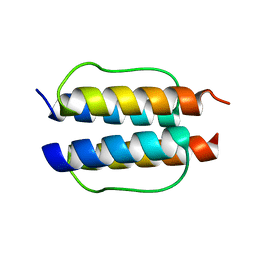 | |
2WPY
 
 | | GCN4 leucine zipper mutant with one VxxNxxx motif coordinating chloride | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Zeth, K, Hartmann, M.D, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WPZ
 
 | | GCN4 leucine zipper mutant with two VxxNxxx motifs coordinating chloride | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Zeth, K, Hartmann, M.D, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XMX
 
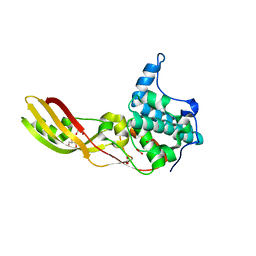 | | High resolution structure of Colicin M | | Descriptor: | CARBONATE ION, COLICIN-M, GLYCEROL | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Activation of Colicin M by the Fkpa Prolyl Cis- Trans Isomerase/Chaperone.
J.Biol.Chem., 286, 2011
|
|
2YMU
 
 | |
2YH5
 
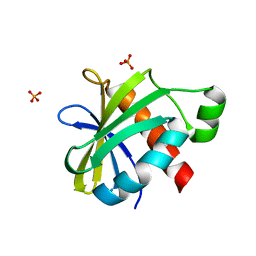 | | Structure of the C-terminal domain of BamC | | Descriptor: | DAPX PROTEIN, PHOSPHATE ION | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-25 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
2YH6
 
 | |
2YHC
 
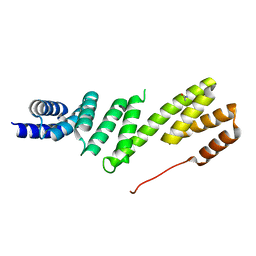 | | Structure of BamD from E. coli | | Descriptor: | UPF0169 LIPOPROTEIN YFIO, UREA | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-01 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
2YH9
 
 | |
2YMK
 
 | |
2YMS
 
 | |
2YJJ
 
 | |
2YJK
 
 | |
1C1Z
 
 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | Authors: | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
