1U6T
 
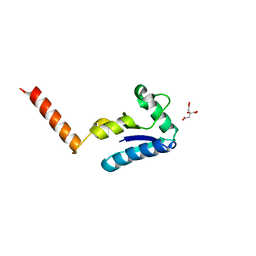 | | Crystal structure of the human SH3 binding glutamic-rich protein like | | Descriptor: | CITRIC ACID, SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Yin, L, Xiang, Y, Yang, N, Zhu, D.-Y, Huang, R.-H, Wang, D.-C. | | Deposit date: | 2004-08-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human SH3BGRL protein: the first structure of the human SH3BGR family representing a novel class of thioredoxin fold proteins
Proteins, 61, 2005
|
|
5XN7
 
 | |
7XGB
 
 | |
6QNS
 
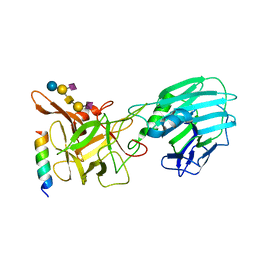 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B mutant I1248W/V1249W in complex with human synaptotagmin 1 and GD1a receptors | | Descriptor: | Botulinum neurotoxin type B, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Synaptotagmin-1 | | Authors: | Masuyer, G, Yin, L, Zhang, S, Miyashita, S.I, Dong, M, Stenmark, P. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy.
Plos Biol., 18, 2020
|
|
3RGV
 
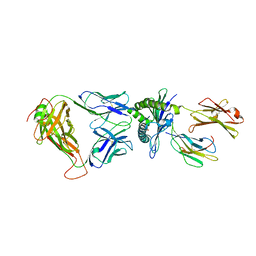 | | A single TCR bound to MHCI and MHC II reveals switchable TCR conformers | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Dai, S, Huseby, E, Scott-Browne, J, Rubtsova, K, Pinilla, C, Crawford, F, Marrack, P, Yin, L, Kappler, J.W. | | Deposit date: | 2011-04-09 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Single T Cell Receptor Bound to Major Histocompatibility Complex Class I and Class II Glycoproteins Reveals Switchable TCR Conformers.
Immunity, 35, 2011
|
|
4H1L
 
 | | TCR interaction with peptide mimics of nickel offers structural insights in nickel contact allergy | | Descriptor: | Ani2.3 TCR A chain, Ani2.3 TCR B chain, HLA class II histocompatibility antigen, ... | | Authors: | Kappler, J.W, Yin, L, Dai, S, Marrack, P. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-14 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T-cell receptor (TCR) interaction with peptides that mimic nickel offers insight into nickel contact allergy.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4H26
 
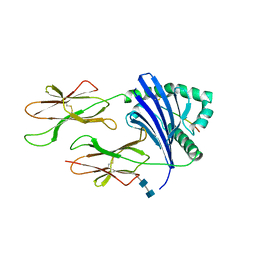 | | TCR interaction with peptide mimics of nickel offers structure insight to nickel contact allergy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Kappler, J.W, Yin, L, Dai, S, Marrack, P, Crawford, F. | | Deposit date: | 2012-09-12 | | Release date: | 2013-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T-cell receptor (TCR) interaction with peptides that mimic nickel offers insight into nickel contact allergy.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HEH
 
 | |
4H25
 
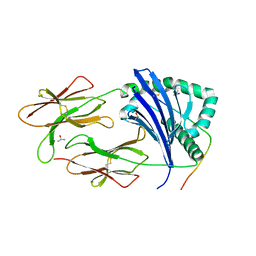 | | TCR interaction with peptide mimics of nickel offers structure insights to nickel contact allergy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Kappler, J.W, Yin, L, Dai, S, Marrack, P, Crawford, F. | | Deposit date: | 2012-09-12 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | T-cell receptor (TCR) interaction with peptides that mimic nickel offers insight into nickel contact allergy.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OV5
 
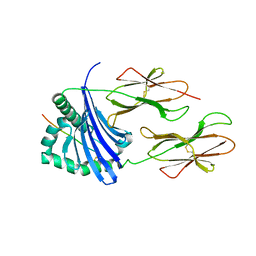 | | Structure of HLA-DR1 with a bound peptide with non-optimal alanine in the P1 pocket | | Descriptor: | HLA class I histocompatibility antigen, A-2 alpha chain, HLA class II histocompatibility antigen, ... | | Authors: | Trenh, P, Yin, L, Stern, L.J. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Susceptibility to HLA-DM Protein Is Determined by a Dynamic Conformation of Major Histocompatibility Complex Class II Molecule Bound with Peptide.
J.Biol.Chem., 289, 2014
|
|
7MC6
 
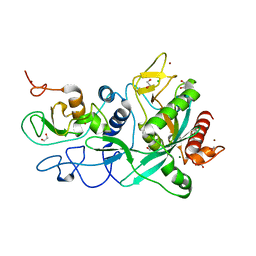 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
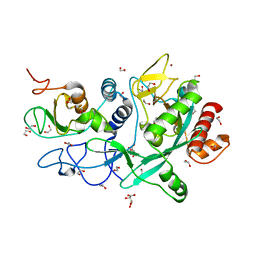 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3NI2
 
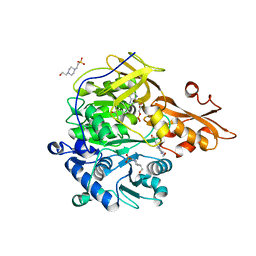 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
7K33
 
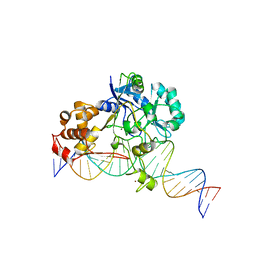 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
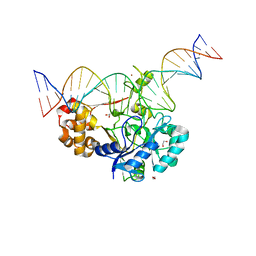 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | Descriptor: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K32
 
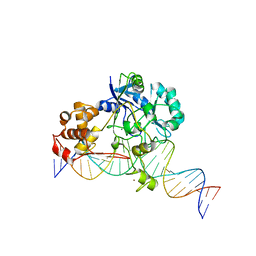 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
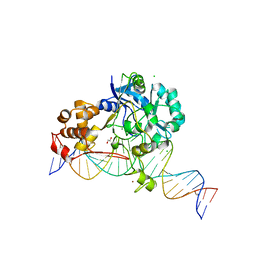 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5YLF
 
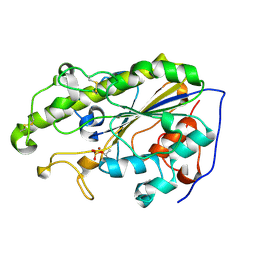 | | MCR-1 complex with D-glucose | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-D-glucopyranose | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
5YLC
 
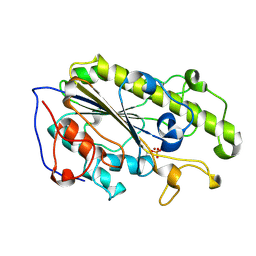 | | Crystal Structure of MCR-1 Catalytic Domain | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
5YLE
 
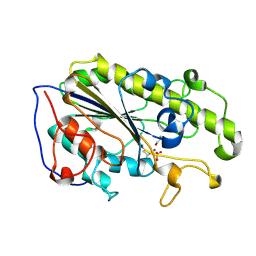 | | MCR-1 complex with ethanolamine (ETA) | | Descriptor: | ETHANOLAMINE, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
6JJ8
 
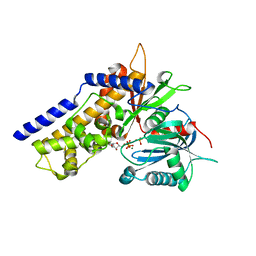 | | Crystal structure of OsHXK6-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of OsHXK6-ATP-Mg2+ complex
To Be Published
|
|
6JJ7
 
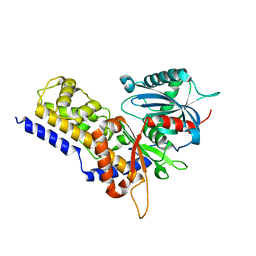 | | Crystal structure of OsHXK6-Glc complex | | Descriptor: | Rice hexokinase 6, beta-D-glucopyranose | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of OsHXK6-Glc complex
To Be Published
|
|
6JQ3
 
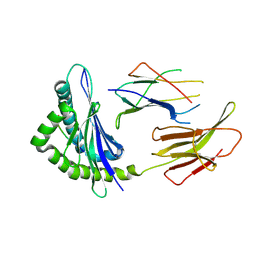 | | Crystal Structure of H2-Kb in complex with a DPAGT1 mutant peptide | | Descriptor: | Beta-2-microglobulin, DPAGT1 mutant antigen SIIVFNLL, H-2 class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Yin, L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JQ2
 
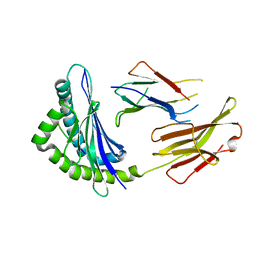 | | Crystal Structure of H2-Kb in complex with a DPAGT1 self-peptide | | Descriptor: | Beta-2-microglobulin, DPATG1 antigen SIIVFNLV, H-2 class I histocompatibility antigen, ... | | Authors: | Bai, P, Yin, L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JJ4
 
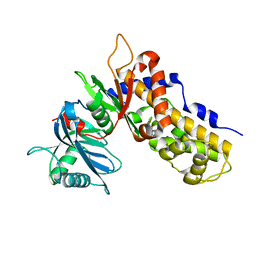 | | Crystal structure of OsHXK6-apo form | | Descriptor: | Hexokinase-6 | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of OsHXK6-apo
To Be Published
|
|
