4OQ4
 
 | | Crystal Structure of E18A Human DJ-1 | | Descriptor: | Protein DJ-1, SODIUM ION | | Authors: | Prahlad, J, Hauser, D.N, Milkovic, N.M, Cookson, M.R, Wilson, M.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of cysteine-reactive cross-linkers to probe conformational flexibility of human DJ-1 demonstrates that Glu18 mutations are dimers.
J Neurochem, 130, 2014
|
|
4PTH
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 100K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PST
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 277 K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PTJ
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 277K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PSS
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 100K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.849 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
3QOU
 
 | | Crystal Structure of E. coli YbbN | | Descriptor: | CALCIUM ION, protein ybbN | | Authors: | Lin, J, Wilson, M.A. | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli Thioredoxin-like Protein YbbN Contains an Atypical Tetratricopeptide Repeat Motif and Is a Negative Regulator of GroEL.
J.Biol.Chem., 286, 2011
|
|
2RGX
 
 | | Crystal Structure of Adenylate Kinase from Aquifex Aeolicus in complex with Ap5A | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Thai, V, Wolf-Watz, M, Fenn, T, Pozharski, E, Wilson, M.A, Petsko, G.A, Kern, D. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intrinsic motions along an enzymatic reaction trajectory.
Nature, 450, 2007
|
|
2RK4
 
 | | Structure of M26I DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
2RH5
 
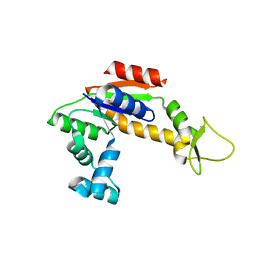 | | Structure of Apo Adenylate Kinase from Aquifex Aeolicus | | Descriptor: | Adenylate kinase | | Authors: | Thai, V, Wolf-Watz, M, Fenn, T, Pozharski, E, Wilson, M.A, Petsko, G.A, Kern, D. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Intrinsic motions along an enzymatic reaction trajectory.
Nature, 450, 2007
|
|
2RK3
 
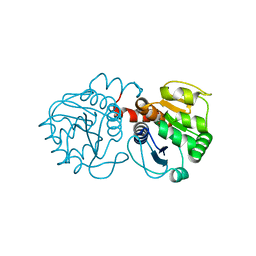 | | Structure of A104T DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
2RK6
 
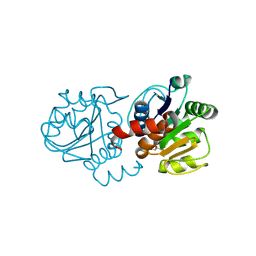 | | Structure of E163K DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
4GE0
 
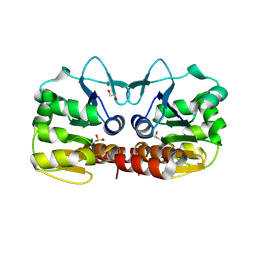 | | Schizosaccharomyces pombe DJ-1 T114P mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
4GE3
 
 | | Schizosaccharomyces pombe DJ-1 T114V mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
4K00
 
 | | Crystal structure of Slr0204, a 1,4-dihydroxy-2-naphthoyl-CoA thioesterase from Synechocystis | | Descriptor: | 1,2-ETHANEDIOL, 1,4-dihydroxy-2-naphthoyl-CoA hydrolase | | Authors: | Furt, F, Allen, W.J, Widhalm, J.R, Madzelan, P, Rizzo, R.C, Basset, G, Wilson, M.A. | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional convergence of structurally distinct thioesterases from cyanobacteria and plants involved in phylloquinone biosynthesis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K02
 
 | | Crystal structure of AtDHNAT1, a 1,4-dihydroxy-2-naphthoyl-CoA thioesterase from Arabidopsis thaliana | | Descriptor: | 1,4-dihydroxy-2-naphthoyl-CoA thioesterase | | Authors: | Furt, F, Allen, W.J, Widhalm, J.R, Madzelan, P, Rizzo, R.C, Basset, G, Wilson, M.A. | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional convergence of structurally distinct thioesterases from cyanobacteria and plants involved in phylloquinone biosynthesis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1OT5
 
 | | The 2.4 Angstrom Crystal Structure of Kex2 in complex with a peptidyl-boronic acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-Ala-Lys-boroArg N-acetylated boronic acid peptide inhibitor, ... | | Authors: | Holyoak, T, Wilson, M.A, Fenn, T.D, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-06-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A Resolution Crystal Structure of the Prototypical Hormone-Processing Protease Kex2 in Complex with an Ala-Lys-Arg Boronic Acid Inhibitor
Biochemistry, 42, 2003
|
|
1Q0X
 
 | | Anti-morphine Antibody 9B1 Unliganded Form | | Descriptor: | Fab 9B1, heavy chain, light chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
1Q0Y
 
 | | Anti-Morphine Antibody 9B1 Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, Fab 9B1, Heavy chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
7LDO
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-3) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9S
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9W
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA0
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LD6
 
 | | G150A Pseudomonas fluorescens isocyanide hydratase (G150A-1) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Q
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Z
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
