7Y0E
 
 | | Crystal structure of TMPRSS2 in complex with Camostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | Descriptor: | Non-structural protein 1 | | Authors: | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
8IQM
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl2 modifying factor, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQK
 
 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl-2-like protein 1, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQL
 
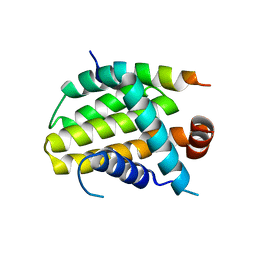 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9577 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
7EOT
 
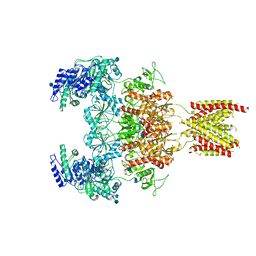 | |
7EOQ
 
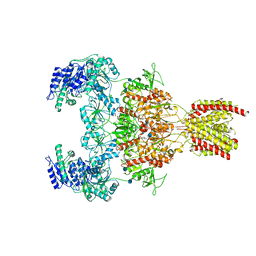 | |
7EOU
 
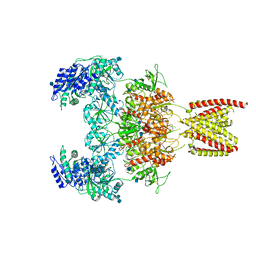 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EOS
 
 | |
7EOR
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
8H1L
 
 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1K
 
 | | Crystal structure of glucose-2-epimerase from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, GLYCEROL, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1M
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7B8V
 
 | |
7B90
 
 | | Circular permutant of ribosomal protein S6, P54-55 truncated, I8A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Circular permutant of ribosomal protein S6, P54-55 truncate, I8A
To Be Published
|
|
7BFG
 
 | |
7BFC
 
 | |
7BFF
 
 | |
7BFE
 
 | |
7BFD
 
 | |
6ISB
 
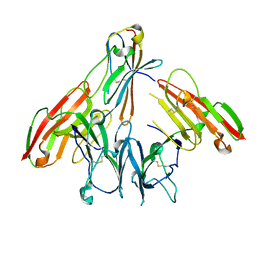 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6LIV
 
 | |
6ISA
 
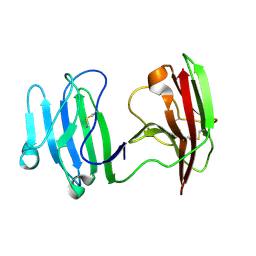 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6LNQ
 
 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | Descriptor: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | Authors: | Wang, H, Shang, L.Q. | | Deposit date: | 2020-01-01 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
