7CIS
 
 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7DHI
 
 | | Cryo-EM structure of the partial agonist salbutamol-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Ling, S.L, Zhou, Y.X, Zhang, Y.N, Lv, P, Liu, S.L, Fang, W, Sun, W.J, Hu, L.Y.A. | | Deposit date: | 2020-11-15 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Different Conformational Responses of the beta2-Adrenergic Receptor-Gs Complex upon Binding of the Partial Agonist Salbutamol or the Full Agonist Isoprenaline
Natl Sci Rev, 2020
|
|
7DHR
 
 | | Cryo-EM structure of the full agonist isoprenaline-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Ling, S.L, Zhou, Y.X, Zhang, Y.N, Lv, P, Liu, S.L, Fang, W, Sun, W.J, Hu, L.Y.A. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Different Conformational Responses of the beta2-Adrenergic Receptor-Gs Complex upon Binding of the Partial Agonist Salbutamol or the Full Agonist Isoprenaline
Natl Sci Rev, 2020
|
|
7DCL
 
 | |
7EDO
 
 | | First insight into marsupial MHC I peptide presentation: immune features of lower mammals paralleled with bats | | Descriptor: | Beta-2-microglobulin, CYS-ASN-VAL-THR-LEU-ASN-TYR-PRO, MHC class I antigen | | Authors: | Wang, P.Y, Yue, C, Lu, D, Liu, K.F, Liu, S, Yao, S.J, Chai, Y, Qi, J.X, Lou, Y.L, Sun, Z.Y, Gao, G.F, Liu, W.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
J Immunol., 207, 2021
|
|
7EMD
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-GLY-ASP-PHE-PHE-HIS-ASP-MET-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EM9
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, SER-LEU-ASP-GLU-TYR-SER-SER-ASP-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMA
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-SER-SER-ASP-VAL-THR-THR-LEU-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMB
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-ALA-ALA-ILE-GLU-GLU-GLU-ASP-ILE, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMC
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-THR-GLU-ILE-ARG-GLU-LEU-LEU-VAL, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7DYN
 
 | | Phosphorylation of MHC I peptide | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
6DHW
 
 | |
6DLU
 
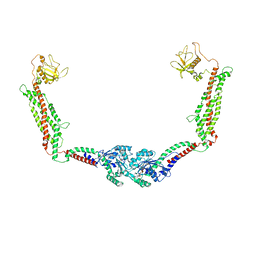 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6DU0
 
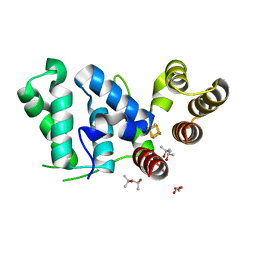 | |
6DI6
 
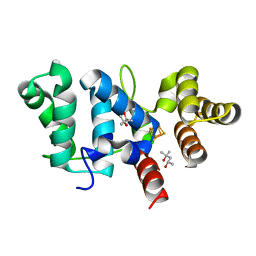 | |
6DTV
 
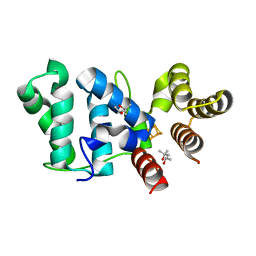 | |
6DE9
 
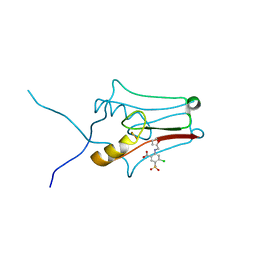 | | mitoNEET bound to furosemide | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Robart, A.R, Geldenhuys, W.J. | | Deposit date: | 2018-05-11 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the mitochondrial protein mitoNEET bound to a benze-sulfonide ligand.
Commun Chem, 2, 2019
|
|
6DI2
 
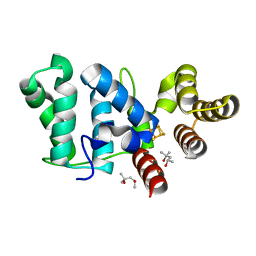 | |
6DLV
 
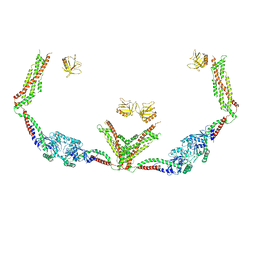 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6DTZ
 
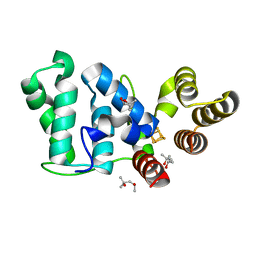 | | Crystal structure of eukaryotic DNA primase large subunit iron-sulfur cluster domain, Y397F mutant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, ... | | Authors: | Salay, L.E, Chazin, W.J. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Yeast require redox switching in DNA primase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E27
 
 | | The CARD9 CARD domain-swapped dimer with a zinc ion bound to one of the two zinc binding sites | | Descriptor: | Caspase recruitment domain-containing protein 9, ZINC ION | | Authors: | Holliday, M.J, Ferrao, R, Boenig, G, Deuber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Picomolar zinc binding modulates formation of Bcl10-nucleating assemblies of the caspase recruitment domain (CARD) of CARD9.
J. Biol. Chem., 293, 2018
|
|
6E28
 
 | | The CARD9 CARD domain-swapped dimer | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Ferrao, R, Boenig, G, Deuber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Picomolar zinc binding modulates formation of Bcl10-nucleating assemblies of the caspase recruitment domain (CARD) of CARD9.
J. Biol. Chem., 293, 2018
|
|
6E26
 
 | |
6E25
 
 | |
