5MJC
 
 | | metNeuroglobin under oxygen at 50 bar | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, OXYGEN MOLECULE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5MNS
 
 | | Structural and functional characterization of OleP in complex with 6DEB in sodium formate | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Parisi, G, Savino, C, Montemiglio, L.C, Vallone, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Substrate-induced conformational change in cytochrome P450 OleP.
FASEB J., 33, 2019
|
|
2VRY
 
 | | Mouse Neuroglobin with heme iron in the reduced ferrous state | | Descriptor: | NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Arcovito, A, Moschetti, T, D'Angelo, P, Mancini, G, Vallone, B, Brunori, M, Della Longa, S. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An X-Ray Diffraction and X-Ray Absorption Spectroscopy Joint Study of Neuroglobin.
Arch.Biochem.Biophys., 475, 2008
|
|
2C1W
 
 | | The structure of XendoU: a splicing independent snoRNA processing endoribonuclease | | Descriptor: | ENDOU PROTEIN, PHOSPHATE ION | | Authors: | Renzi, F, Caffarelli, E, Laneve, P, Bozzoni, I, Brunori, M, Vallone, B. | | Deposit date: | 2005-09-21 | | Release date: | 2006-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Endoribonuclease Xendou: From Small Nucleolar RNA Processing to Severe Acute Respiratory Syndrome Coronavirus Replication.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2C9J
 
 | | Structure of the fluorescent protein cmFP512 at 1.35A from Cerianthus membranaceus | | Descriptor: | GREEN FLUORESCENT PROTEIN FP512 | | Authors: | Renzi, F, Nienhaus, K, Wiedenmann, J, Vallone, B, Nienhaus, G.U. | | Deposit date: | 2005-12-12 | | Release date: | 2006-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploring Chromophore-Protein Interactions in Fluorescent Protein Cmfp512 from Cerianthus Membranaceus: X-Ray Structure Analysis and Optical Spectroscopy.
Biochemistry, 45, 2006
|
|
2C9I
 
 | | Structure of the fluorescent protein asFP499 from Anemonia sulcata | | Descriptor: | GREEN FLUORESCENT PROTEIN ASFP499 | | Authors: | Renzi, F, Nienhaus, K, Wiedenmann, J, Vallone, B, Nienhaus, G.U. | | Deposit date: | 2005-12-12 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chromophore-Protein Interactions in the Anthozoan Green Fluorescent Protein Asfp499
Biophys.J., 91, 2006
|
|
1QI8
 
 | | DEOXYGENATED STRUCTURE OF A DISTAL POCKET HEMOGLOBIN MUTANT | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miele, A.E, Vallone, B, Santanche, S, Travaglini-Allocatelli, C, Bellelli, A, Brunori, M. | | Deposit date: | 1999-06-07 | | Release date: | 1999-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of ligand binding in engineered human hemoglobin distal pocket.
J.Mol.Biol., 290, 1999
|
|
3GK9
 
 | | Crystal structure of murine Ngb under Xe pressure | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
3GLN
 
 | | Carbonmonoxy Ngb under Xenon pressure | | Descriptor: | CARBON MONOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
3GKT
 
 | | Crystal structure of murine neuroglobin under Kr pressure | | Descriptor: | KRYPTON, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
1MYZ
 
 | | CO COMPLEX OF MYOGLOBIN MB-YQR AT RT SOLVED FROM LAUE DATA. | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bourgeois, D, Vallone, B, Schotte, F, Arcovito, A, Miele, A.E, Sciara, G, Wulff, M, Anfinrud, P, Brunori, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex landscape of protein
structural dynamics unveiled by
nanosecond Laue crystallography.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MZ0
 
 | | STRUCTURE OF MYOGLOBIN MB-YQR 316 ns AFTER PHOTOLYSIS OF CARBON MONOXIDE SOLVED FROM LAUE DATA AT RT. | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bourgeois, D, Vallone, B, Schotte, F, Arcovito, A, Miele, A.E, Sciara, G, Wulff, M, Anfinrud, P, Brunori, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex landscape of protein structural dynamics unveiled by
nanosecond Laue crystallography.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7OHD
 
 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
1J7S
 
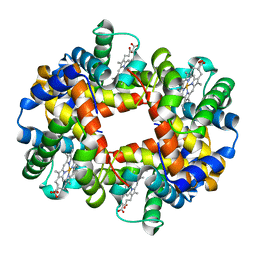 | | Crystal Structure of deoxy HbalphaYQ, a mutant of HbA | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
1J7W
 
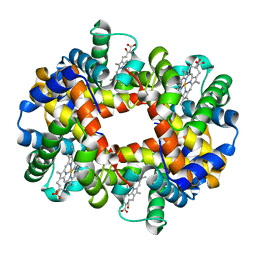 | | Crystal structure of deoxy HbbetaYQ, a site directed mutant of HbA | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
6XUV
 
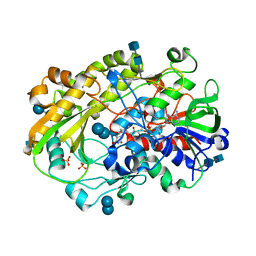 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, laminaribiose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6XUU
 
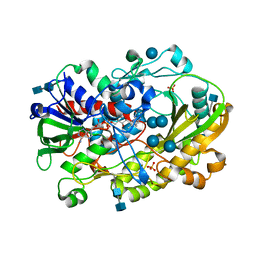 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, glucose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6XUT
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Sciara, G, Vallone, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6ZI7
 
 | | Crystal structure of OleP-oleandolide(DEO) bound to L-rhamnose | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZHZ
 
 | | OleP-oleandolide(DEO) in high salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI3
 
 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI2
 
 | | OleP-oleandolide(DEO) in low salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
5N9X
 
 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, ligand bound structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylation domain, MAGNESIUM ION, ... | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
5N9W
 
 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, apo structure | | Descriptor: | ACETATE ION, Adenylation domain | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
5O17
 
 | | Crystal structure of murine neuroglobin under 100 bar krypton | | Descriptor: | KRYPTON, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Colloc'h, N, Prange, T, Vallone, B, Carpentier, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping Hydrophobic Tunnels and Cavities in Neuroglobin with Noble Gas under Pressure.
Biophys. J., 113, 2017
|
|
