3N04
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | GLYCEROL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
3NJA
 
 | | The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472. | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable GGDEF family protein, ... | | Authors: | Tan, K, Wu, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472.
To be Published
|
|
3NKZ
 
 | | The crystal structure of a flagella protein from Yersinia enterocolitica subsp. enterocolitica 8081 | | Descriptor: | Flagellar protein fliT, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Li, H, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | The crystal structure of a flagella protein from Yersinia enterocolitica subsp. enterocolitica 8081
To be Published
|
|
3NUK
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | ALPHA-GLUCOSIDASE, GLYCEROL | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174
TO BE PUBLISHED
|
|
3NSX
 
 | | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174
To be Published
|
|
3OCM
 
 | | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative membrane protein, ... | | Authors: | Tan, K, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis
To be Published
|
|
3OCO
 
 | |
3OD1
 
 | | The crystal structure of an ATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C | | Descriptor: | ATP phosphoribosyltransferase regulatory subunit, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Bigelow, L, Hamilton, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of aATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C
To be Published
|
|
3OLQ
 
 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
3Q13
 
 | | The Structure of the Ca2+-binding, Glycosylated F-spondin Domain of F-spondin, A C2-domain Variant from Extracellular Matrix | | Descriptor: | ACETATE ION, CALCIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Lawler, J. | | Deposit date: | 2010-12-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the Ca2+-binding, glycosylated F-spondin domain of F-spondin - A C2-domain variant in an extracellular matrix protein.
Bmc Struct.Biol., 11, 2011
|
|
3PNN
 
 | |
3PLX
 
 | | The crystal structure of aspartate alpha-decarboxylase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, Aspartate 1-decarboxylase, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | The crystal structure of aspartate alpha-decarboxylase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3POC
 
 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
3PMM
 
 | | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | FORMIC ACID, IMIDAZOLE, Putative cytoplasmic protein | | Authors: | Tan, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-29 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3PN8
 
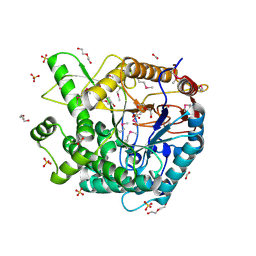 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Putative phospho-beta-glucosidase, ... | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-15 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutants UA159
To be Published
|
|
3PHA
 
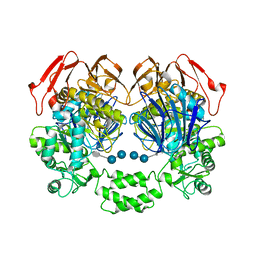 | | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose
To be Published
|
|
3QWT
 
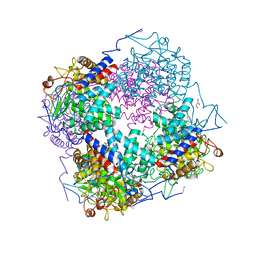 | |
3QAO
 
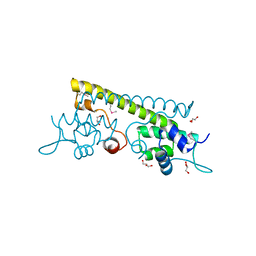 | | The crystal structure of the N-terminal domain of a MerR-like transcriptional regulator from Listeria monocytogenes EGD-e | | Descriptor: | GLYCEROL, MerR-like transcriptional regulator | | Authors: | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-11 | | Release date: | 2011-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | The crystal structure of the N-terminal domain of a MerR-like transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
3QZ3
 
 | | The crystal structure of ferritin from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Ferritin | | Authors: | Tan, K, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The crystal structure of ferritin from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3QZ6
 
 | |
3RPW
 
 | | The crystal structure of an ABC transporter from Rhodopseudomonas palustris CGA009 | | Descriptor: | ABC transporter, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3ROB
 
 | |
3RF1
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Glycyl-tRNA synthetase alpha subunit | | Authors: | Tan, K, Zhang, R, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-05 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3R0V
 
 | | The crystal structure of an alpha/beta hydrolase from Sphaerobacter thermophilus DSM 20745. | | Descriptor: | Alpha/beta hydrolase fold protein, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | The crystal structure of an alpha/beta hydrolase from Sphaerobacter thermophilus DSM 20745.
To be Published
|
|
3RPC
 
 | |
