1OFD
 
 | |
1OFE
 
 | |
4LZ2
 
 | | Crystal structure of the bromodomain of human BAZ2A | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION | | Authors: | Tallant, C, Nunez-Alonso, G, Picaud, S, Filippakopoulos, P, Krojer, T, Bradley, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4MB4
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB5
 
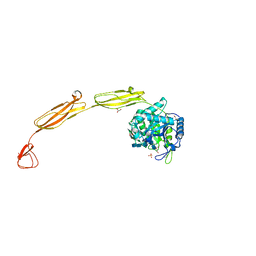 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
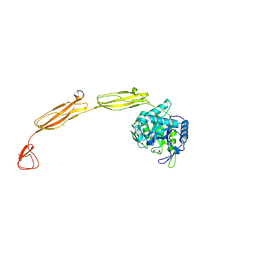 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NK7
 
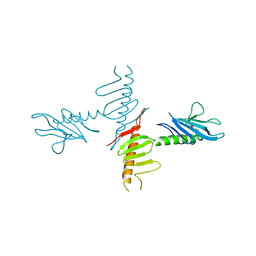 | |
4NKB
 
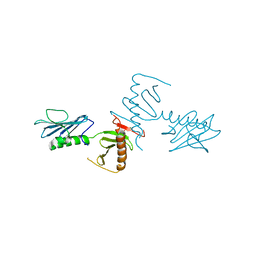 | | Crystal Structure of the cryptic polo box (CPB)of ZYG-1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Shimanovskaya, E, Dong, G. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C. elegans ZYG-1 Cryptic Polo Box Suggests a Conserved Mechanism for Centriolar Docking of Plk4 Kinases.
Structure, 22, 2014
|
|
4OW2
 
 | | YopM from Yersinia enterocolitica WA-314 | | Descriptor: | Yop effector YopM | | Authors: | Rumm, A, Perbandt, M, Aepfelbacher, M. | | Deposit date: | 2014-01-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog., 12, 2016
|
|
4QC3
 
 | | Crystal structure of human BAZ2B bromodomain in complex with a diacetylated histone 4 peptide (H4K8acK12ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, diacetylated histone 4 peptide (H4K8acK12ac) | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4QC1
 
 | | Crystal structure of human BAZ2B bromodomain in complex with an acetylated histone 3 peptide (H3K14ac) | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, SULFATE ION, ZINC ION, ... | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
2O0T
 
 | |
2OQR
 
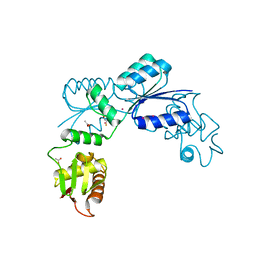 | |
2PUW
 
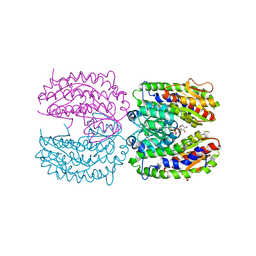 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, isomerase domain of glutamine-fructose-6-phosphate transaminase (isomerizing) | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
2PUV
 
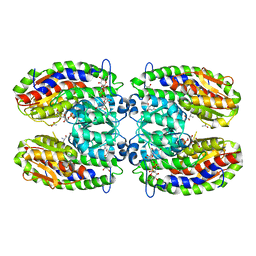 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 5-AMINO-5-DEOXY-1-O-PHOSPHONO-D-MANNITOL, ACETATE ION, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
2POC
 
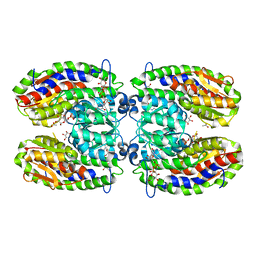 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETATE ION, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
2PUT
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | ACETATE ION, FRUCTOSE -6-PHOSPHATE, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
2QKL
 
 | |
2QKM
 
 | |
2R15
 
 | |
2QEQ
 
 | |
2RA1
 
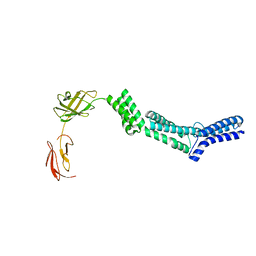 | | Crystal structure of the N-terminal part of the bacterial S-layer protein SbsC | | Descriptor: | Surface layer protein | | Authors: | Pavkov, T, Egelseer, E.M, Tesarz, M, Sleytr, U.B, Keller, W. | | Deposit date: | 2007-09-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | The structure and binding behavior of the bacterial cell surface layer protein SbsC.
Structure, 16, 2008
|
|
3LPW
 
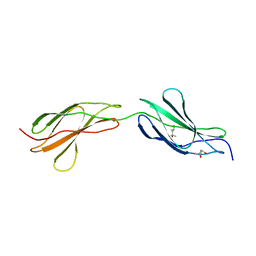 | | Crystal structure of the FnIII-tandem A77-A78 from the A-band of titin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, A77-A78 domain from Titin | | Authors: | Bucher, R.M, Mayans, O. | | Deposit date: | 2010-02-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the FnIII Tandem A77-A78 points to a periodically conserved architecture in the myosin-binding region of titin
J.Mol.Biol., 401, 2010
|
|
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O1B
 
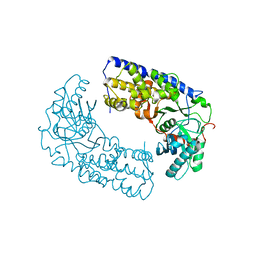 | | CRYSTAL STRUCTURE OF DIMERIC KLHXK1 IN CRYSTAL FORM II | | Descriptor: | Hexokinase | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-21 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
